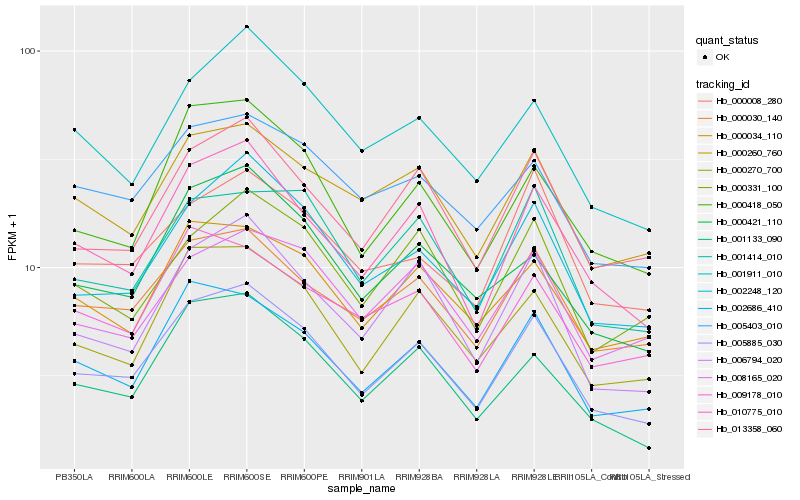
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005885_030 |
0.0 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 2 |
Hb_010775_010 |
0.0531254426 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002686_410 |
0.0633392299 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000270_700 |
0.0672452585 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 5 |
Hb_002248_120 |
0.0682093351 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 6 |
Hb_006794_020 |
0.06988831 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 7 |
Hb_001133_090 |
0.0713637816 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 8 |
Hb_013358_060 |
0.0715095335 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 9 |
Hb_000030_140 |
0.0734180364 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000418_050 |
0.074453551 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 11 |
Hb_000260_760 |
0.0838080712 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 12 |
Hb_001911_010 |
0.085063849 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 13 |
Hb_000034_110 |
0.0854355904 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 14 |
Hb_000421_110 |
0.086001715 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_005403_010 |
0.0869559891 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 16 |
Hb_001414_010 |
0.0886552767 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 17 |
Hb_000331_100 |
0.0907715779 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000008_280 |
0.0931970585 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 19 |
Hb_009178_010 |
0.0937469015 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 20 |
Hb_008165_020 |
0.0938411947 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |