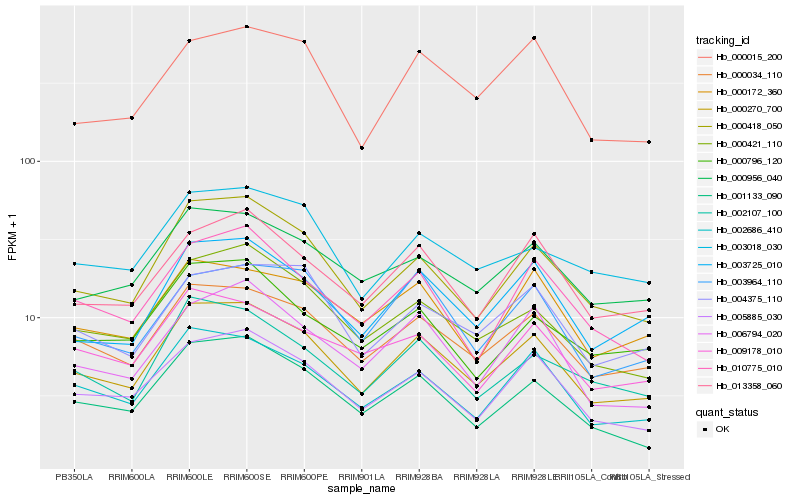
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_700 |
0.0 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 2 |
Hb_000034_110 |
0.0539312274 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 3 |
Hb_000418_050 |
0.0586935068 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 4 |
Hb_002686_410 |
0.0637666097 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_005885_030 |
0.0672452585 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 6 |
Hb_003725_010 |
0.0711444009 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 7 |
Hb_000421_110 |
0.0717490459 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001133_090 |
0.0724089287 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 9 |
Hb_013358_060 |
0.0785189818 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 10 |
Hb_000956_040 |
0.0793945331 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 11 |
Hb_002107_100 |
0.0797805985 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 12 |
Hb_010775_010 |
0.0851509753 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 13 |
Hb_006794_020 |
0.0857363981 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 14 |
Hb_000015_200 |
0.0874701223 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: heat shock cognate 70 kDa protein 2-like [Jatropha curcas] |
| 15 |
Hb_004375_110 |
0.0884125967 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 16 |
Hb_003018_030 |
0.0894433593 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 17 |
Hb_009178_010 |
0.0895177747 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 18 |
Hb_003964_110 |
0.0911118945 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000796_120 |
0.0921564448 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000172_360 |
0.0922682218 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |