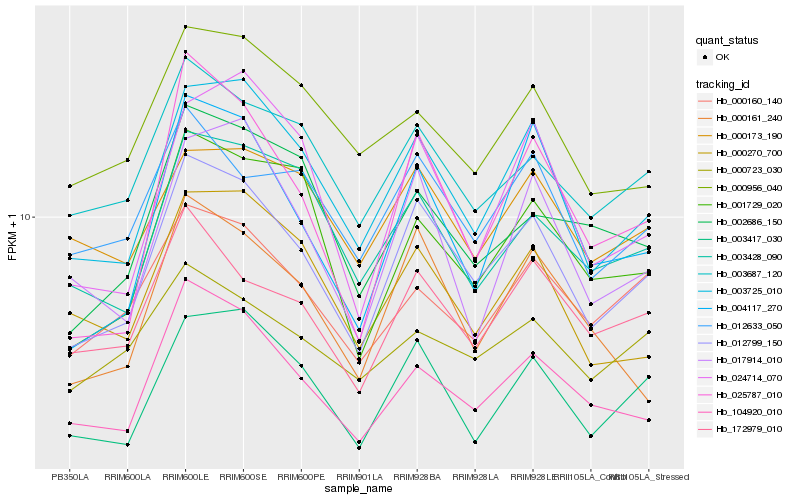
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012799_150 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: methyl-CpG-binding domain-containing protein 13 [Jatropha curcas] |
| 2 |
Hb_003725_010 |
0.0752527104 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 3 |
Hb_172979_010 |
0.098669253 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 4 |
Hb_104920_010 |
0.0994100731 |
- |
- |
hypothetical protein VITISV_031859 [Vitis vinifera] |
| 5 |
Hb_003417_030 |
0.1029554328 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003687_120 |
0.103000391 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 7 |
Hb_025787_010 |
0.1035531978 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 8 |
Hb_004117_270 |
0.1053258666 |
- |
- |
- |
| 9 |
Hb_012633_050 |
0.1087286774 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_003428_090 |
0.110575381 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 11 |
Hb_001729_020 |
0.1108483035 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_024714_070 |
0.1116457201 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 13 |
Hb_000160_140 |
0.112654645 |
- |
- |
PREDICTED: ELL-associated factor 2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000161_240 |
0.1140881805 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_017914_010 |
0.1141300034 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 16 |
Hb_000270_700 |
0.1142244561 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 17 |
Hb_000723_030 |
0.1194026543 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X1 [Jatropha curcas] |
| 18 |
Hb_000173_190 |
0.1216297687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000956_040 |
0.1217500169 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 20 |
Hb_002686_150 |
0.1220567472 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |