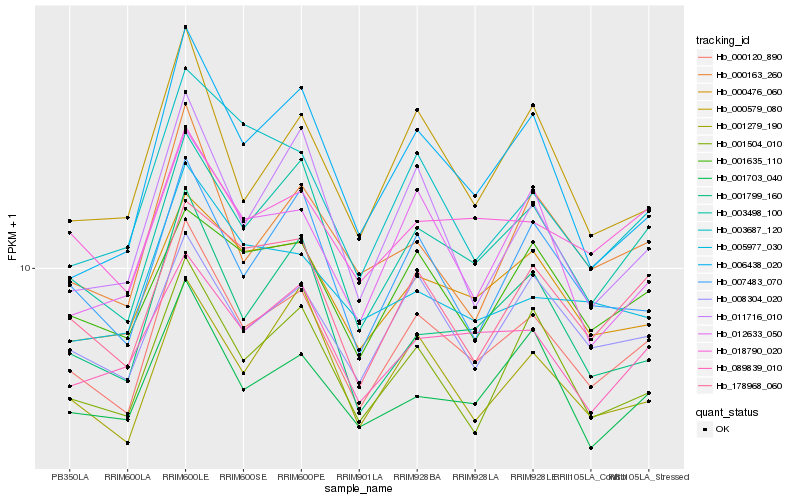
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_890 |
0.0 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 2 |
Hb_003498_100 |
0.0474396837 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 3 |
Hb_000476_060 |
0.0694414077 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_001504_010 |
0.0696122782 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 5 |
Hb_178968_060 |
0.0701998019 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 6 |
Hb_008304_020 |
0.0705009767 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 7 |
Hb_018790_020 |
0.0735662394 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 8 |
Hb_007483_070 |
0.0737389322 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 9 |
Hb_001279_190 |
0.0742804271 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001703_040 |
0.0747132675 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001635_110 |
0.0759572728 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 12 |
Hb_089839_010 |
0.0773587083 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 13 |
Hb_000579_080 |
0.0777129451 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 14 |
Hb_000163_260 |
0.0778157798 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 15 |
Hb_001799_160 |
0.0781377866 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_012633_050 |
0.0825375215 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_006438_020 |
0.084123415 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 18 |
Hb_003687_120 |
0.0841311102 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 19 |
Hb_005977_030 |
0.0858146748 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 20 |
Hb_011716_010 |
0.0859633564 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |