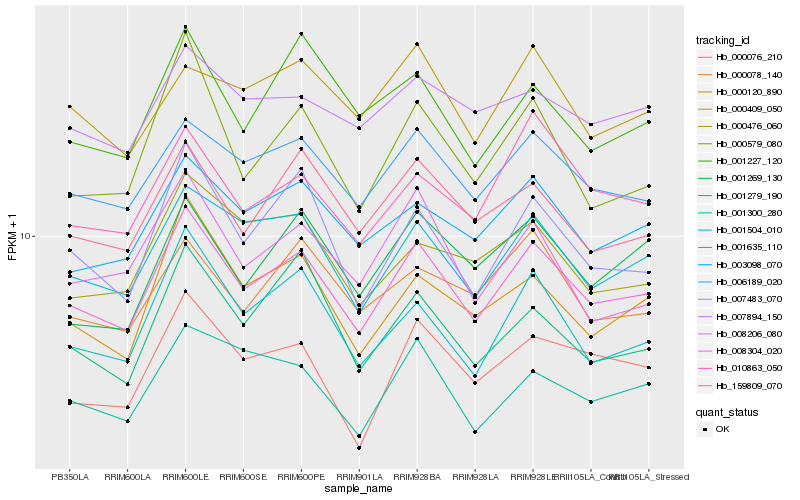
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008304_020 |
0.0 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 2 |
Hb_001635_110 |
0.0536437042 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 3 |
Hb_006189_020 |
0.056093641 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 4 |
Hb_003098_070 |
0.0639183231 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 5 |
Hb_007483_070 |
0.0671526658 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 6 |
Hb_001504_010 |
0.068114396 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 7 |
Hb_159809_070 |
0.0687993854 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 8 |
Hb_000120_890 |
0.0705009767 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 9 |
Hb_000579_080 |
0.0744278765 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 10 |
Hb_010863_050 |
0.0748266831 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 11 |
Hb_000076_210 |
0.0770527743 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit alpha, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_000078_140 |
0.0779271254 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 13 |
Hb_001269_130 |
0.0780570361 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 14 |
Hb_001227_120 |
0.0786226701 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 15 |
Hb_007894_150 |
0.0788984323 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 16 |
Hb_000476_060 |
0.0794600177 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_001300_280 |
0.0798971835 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein ucpB [Jatropha curcas] |
| 18 |
Hb_008206_080 |
0.0799753469 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 17 [Jatropha curcas] |
| 19 |
Hb_001279_190 |
0.0805094278 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000409_050 |
0.0811755227 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |