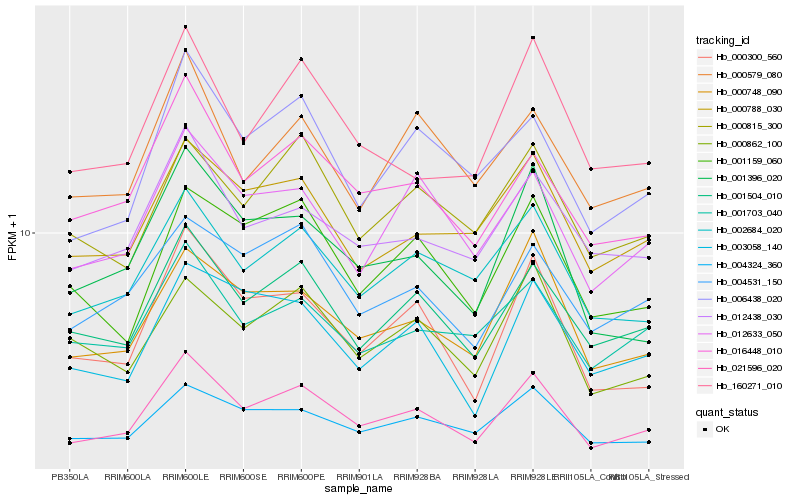
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021596_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 2 |
Hb_001504_010 |
0.0610650054 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 3 |
Hb_002684_020 |
0.0684904179 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 4 |
Hb_016448_010 |
0.0738181069 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 5 |
Hb_000748_090 |
0.0744782727 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 6 |
Hb_006438_020 |
0.0770037922 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 7 |
Hb_012633_050 |
0.0782985438 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_012438_030 |
0.0784953752 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 9 |
Hb_000300_560 |
0.0800487641 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000788_030 |
0.0807301814 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 11 |
Hb_004531_150 |
0.0819821656 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001703_040 |
0.0848355698 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004324_360 |
0.084964007 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001396_020 |
0.0857093141 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 15 |
Hb_000815_300 |
0.0862356944 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 16 |
Hb_001159_060 |
0.0872956083 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 17 |
Hb_000579_080 |
0.0887496356 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 18 |
Hb_000862_100 |
0.0893428913 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003058_140 |
0.0893778195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_160271_010 |
0.0895312165 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |