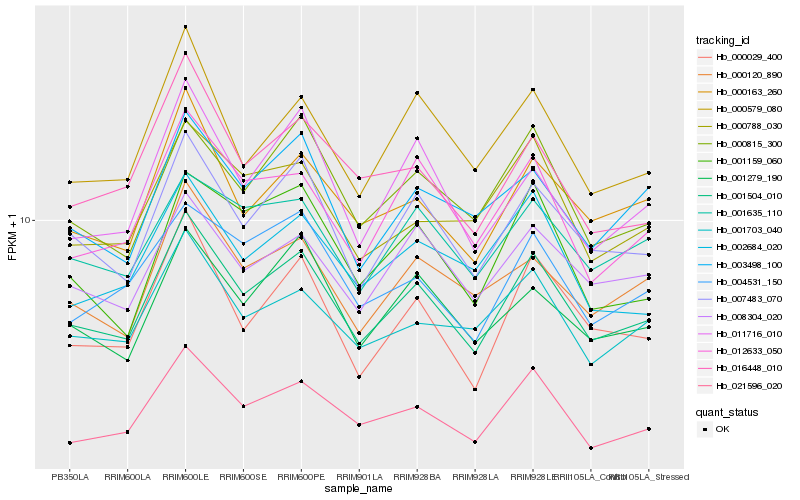
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001504_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 2 |
Hb_007483_070 |
0.0480644372 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 3 |
Hb_021596_020 |
0.0610650054 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 4 |
Hb_000163_260 |
0.0632376045 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 5 |
Hb_000579_080 |
0.0668650036 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 6 |
Hb_008304_020 |
0.068114396 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 7 |
Hb_000815_300 |
0.0683886798 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 8 |
Hb_012633_050 |
0.068614691 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_001279_190 |
0.0692847692 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000029_400 |
0.0693400043 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000120_890 |
0.0696122782 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 12 |
Hb_001703_040 |
0.0713549329 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001635_110 |
0.0733483181 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 14 |
Hb_003498_100 |
0.0749152171 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 15 |
Hb_011716_010 |
0.0749492335 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 16 |
Hb_004531_150 |
0.0754313035 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_016448_010 |
0.0758231462 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 18 |
Hb_002684_020 |
0.0770367937 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 19 |
Hb_000788_030 |
0.0780217477 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 20 |
Hb_001159_060 |
0.0800559871 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |