| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
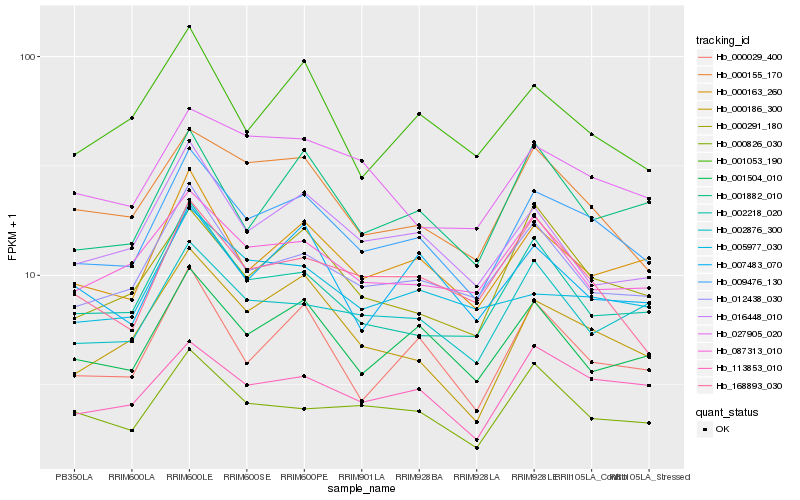
Hb_009476_130 |
0.0 |
- |
- |
hypothetical protein VITISV_005430 [Vitis vinifera] |
| 2 |
Hb_000163_260 |
0.0744143619 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 3 |
Hb_113853_010 |
0.0790475403 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 4 |
Hb_012438_030 |
0.0805203328 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 5 |
Hb_000186_300 |
0.0845000415 |
- |
- |
PREDICTED: malonyl-CoA decarboxylase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001504_010 |
0.0851064137 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 7 |
Hb_000155_170 |
0.0860737012 |
transcription factor |
TF Family: VOZ |
hypothetical protein EUTSA_v10007498mg [Eutrema salsugineum] |
| 8 |
Hb_087313_010 |
0.0907192514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000291_180 |
0.0916256171 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 10 |
Hb_002218_020 |
0.0922101435 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 11 |
Hb_000029_400 |
0.0922636966 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000826_030 |
0.0941254839 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase isoform X1 [Jatropha curcas] |
| 13 |
Hb_001882_010 |
0.0944265122 |
- |
- |
- |
| 14 |
Hb_002876_300 |
0.0945142519 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_016448_010 |
0.0969115421 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 16 |
Hb_168893_030 |
0.0974092514 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 17 |
Hb_027905_020 |
0.0975400909 |
- |
- |
PREDICTED: uncharacterized protein LOC105642268 [Jatropha curcas] |
| 18 |
Hb_001053_190 |
0.0977434755 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 19 |
Hb_007483_070 |
0.098514185 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 20 |
Hb_005977_030 |
0.0994463405 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |