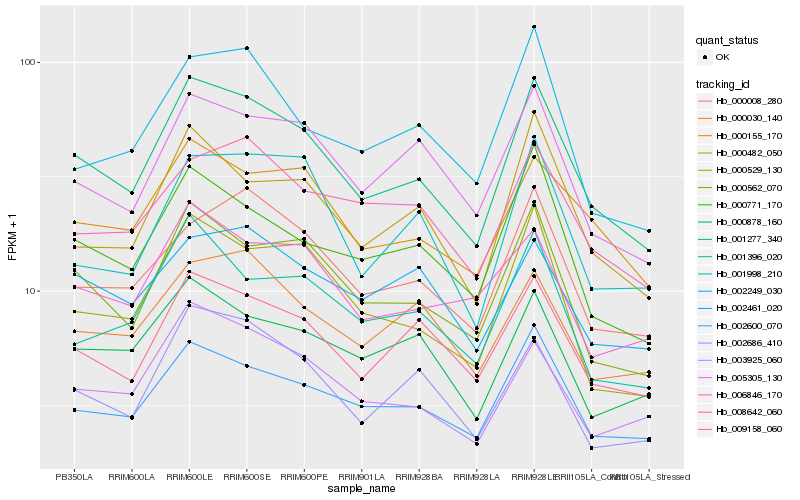
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_340 |
0.0 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 2 |
Hb_000562_070 |
0.069818908 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002600_070 |
0.0711752829 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 4 |
Hb_009158_060 |
0.0770141693 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 5 |
Hb_005305_130 |
0.0833095593 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 6 |
Hb_000155_170 |
0.0833763695 |
transcription factor |
TF Family: VOZ |
hypothetical protein EUTSA_v10007498mg [Eutrema salsugineum] |
| 7 |
Hb_003925_060 |
0.0846865287 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000771_170 |
0.085082508 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 9 |
Hb_000482_050 |
0.0851877913 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000008_280 |
0.0858975586 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 11 |
Hb_000529_130 |
0.0865958755 |
- |
- |
hypothetical protein POPTR_0007s14320g [Populus trichocarpa] |
| 12 |
Hb_002686_410 |
0.0882565327 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001396_020 |
0.0958001088 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 14 |
Hb_000878_160 |
0.0958134935 |
- |
- |
PREDICTED: D-lactate dehydrogenase [cytochrome], mitochondrial [Jatropha curcas] |
| 15 |
Hb_000030_140 |
0.0964369505 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_008642_060 |
0.0974787634 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 17 |
Hb_006846_170 |
0.0994750139 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 18 |
Hb_002249_030 |
0.1001125892 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002461_020 |
0.1018932124 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001998_210 |
0.1024348384 |
- |
- |
unnamed protein product [Coffea canephora] |