| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
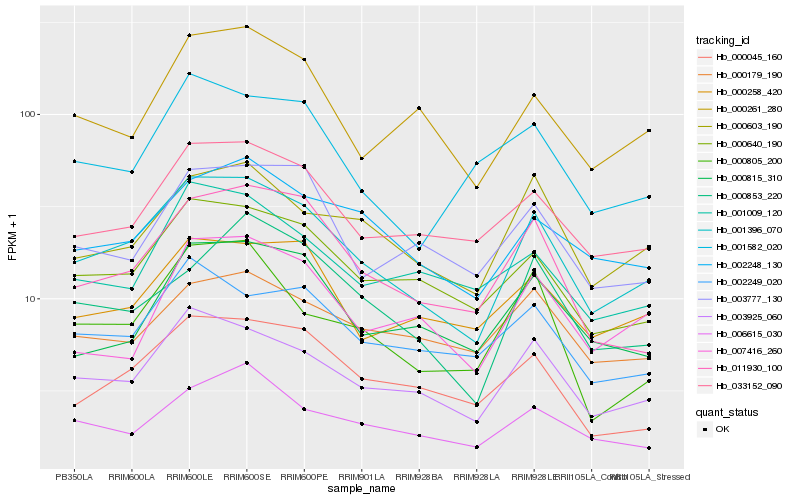
Hb_001396_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003925_060 |
0.0752176019 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000640_190 |
0.0955895393 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_033152_090 |
0.0999947814 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 5 |
Hb_000603_190 |
0.1036986994 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 6 |
Hb_011930_100 |
0.1040508106 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 7 |
Hb_002248_130 |
0.1063165317 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 8 |
Hb_000045_160 |
0.1089611703 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 9 |
Hb_000261_280 |
0.1131723569 |
- |
- |
BnaA10g23810D [Brassica napus] |
| 10 |
Hb_000258_420 |
0.1140200984 |
- |
- |
sucrose transporter 2A [Hevea brasiliensis] |
| 11 |
Hb_000853_220 |
0.1161375695 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 12 |
Hb_000805_200 |
0.1163933706 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002249_020 |
0.1166872581 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 14 |
Hb_000815_310 |
0.116746416 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 15 |
Hb_006615_030 |
0.1171798663 |
- |
- |
prolyl oligopeptidase family protein [Populus trichocarpa] |
| 16 |
Hb_003777_130 |
0.1198924421 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 17 |
Hb_007416_260 |
0.1234155358 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF12-A-like [Jatropha curcas] |
| 18 |
Hb_000179_190 |
0.1235168024 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 19 |
Hb_001582_020 |
0.1241251833 |
- |
- |
plastid acyl-ACP thioesterase [Vernicia fordii] |
| 20 |
Hb_001009_120 |
0.1249972757 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |