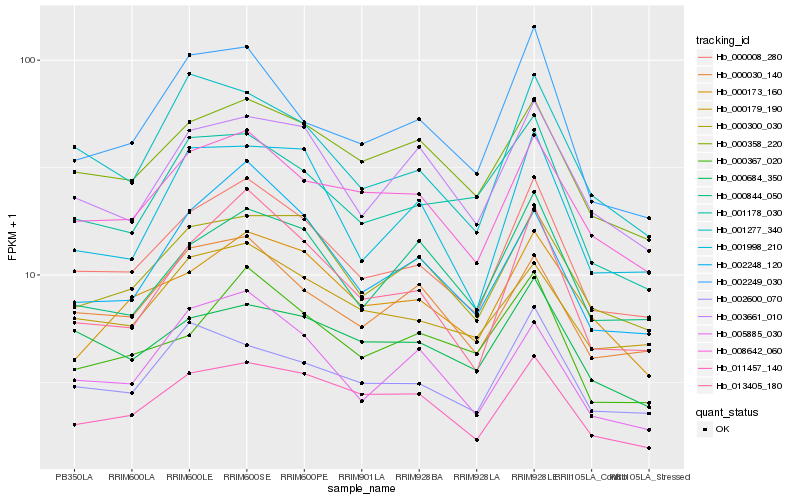
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_280 |
0.0 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 2 |
Hb_008642_060 |
0.0690748503 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 3 |
Hb_013405_180 |
0.0744552857 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 4 |
Hb_000300_030 |
0.0777780314 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 5 |
Hb_000358_220 |
0.0791251412 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 6 |
Hb_000179_190 |
0.080682486 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 7 |
Hb_011457_140 |
0.0823509117 |
- |
- |
PREDICTED: uncharacterized protein LOC105649947 [Jatropha curcas] |
| 8 |
Hb_000030_140 |
0.0827315954 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003661_010 |
0.0841724884 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002249_030 |
0.0850507653 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001277_340 |
0.0858975586 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 12 |
Hb_000844_050 |
0.0882282507 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 13 |
Hb_000367_020 |
0.0889801211 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 14 |
Hb_001178_030 |
0.0898579684 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |
| 15 |
Hb_002248_120 |
0.0910227823 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 16 |
Hb_002600_070 |
0.0914172071 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 17 |
Hb_000173_160 |
0.0919429864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001998_210 |
0.0919819638 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_005885_030 |
0.0931970585 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 20 |
Hb_000684_350 |
0.0951927019 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |