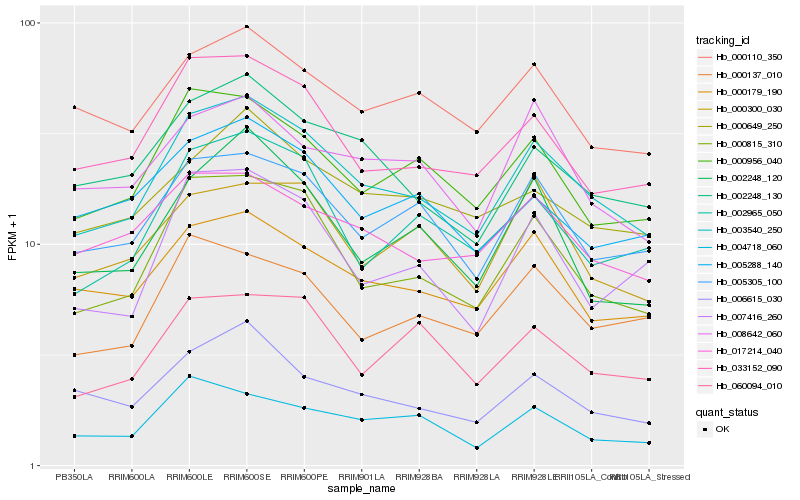
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003540_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000815_310 |
0.0651988555 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 3 |
Hb_005305_100 |
0.0769064707 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_033152_090 |
0.0772354195 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 5 |
Hb_000956_040 |
0.0798682566 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 6 |
Hb_000179_190 |
0.0800787986 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 7 |
Hb_002248_130 |
0.0815820816 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 8 |
Hb_002965_050 |
0.0894726357 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 9 |
Hb_017214_040 |
0.090803255 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase COP1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000137_010 |
0.0911698841 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 11 |
Hb_000110_350 |
0.0939844757 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 12 |
Hb_000649_250 |
0.0943607667 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 13 |
Hb_006615_030 |
0.0944763666 |
- |
- |
prolyl oligopeptidase family protein [Populus trichocarpa] |
| 14 |
Hb_002248_120 |
0.0969510141 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 15 |
Hb_060094_010 |
0.0971456463 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007416_260 |
0.0978061518 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF12-A-like [Jatropha curcas] |
| 17 |
Hb_008642_060 |
0.0978484845 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 18 |
Hb_005288_140 |
0.0989069679 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 19 |
Hb_000300_030 |
0.0998302936 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 20 |
Hb_004718_060 |
0.1031383538 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |