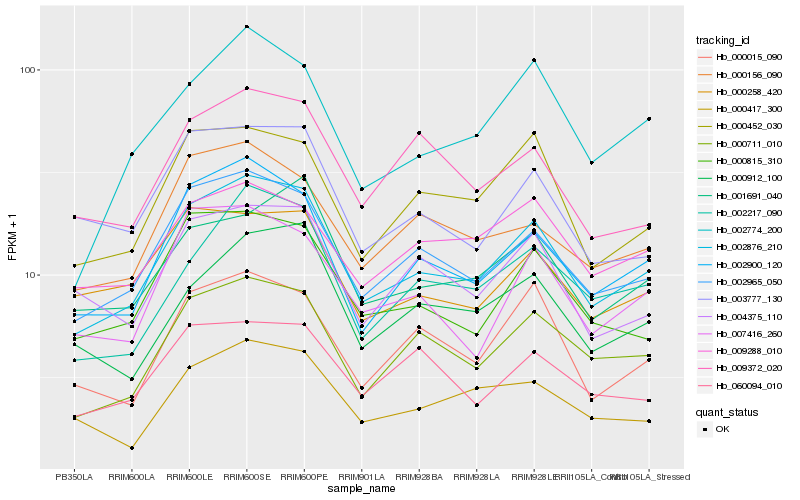
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_210 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 2 |
Hb_002965_050 |
0.0551385609 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 3 |
Hb_002900_120 |
0.0722458619 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 4 |
Hb_000711_010 |
0.0726284499 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 5 |
Hb_000815_310 |
0.0847704443 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000452_030 |
0.0861875129 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 7 |
Hb_002217_090 |
0.0915028089 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 8 |
Hb_000156_090 |
0.0920701637 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007416_260 |
0.0929750961 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF12-A-like [Jatropha curcas] |
| 10 |
Hb_000015_090 |
0.0934972455 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 11 |
Hb_001691_040 |
0.0935805048 |
- |
- |
PREDICTED: uncharacterized protein LOC105649546 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002774_200 |
0.0958251534 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 13 |
Hb_000912_100 |
0.0962458745 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 14 |
Hb_004375_110 |
0.0980291077 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 15 |
Hb_000258_420 |
0.0984623966 |
- |
- |
sucrose transporter 2A [Hevea brasiliensis] |
| 16 |
Hb_000417_300 |
0.1008781083 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7 [Jatropha curcas] |
| 17 |
Hb_003777_130 |
0.1008919282 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 18 |
Hb_060094_010 |
0.1021296749 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 19 |
Hb_009372_020 |
0.1035384628 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 20 |
Hb_009288_010 |
0.1042961552 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |