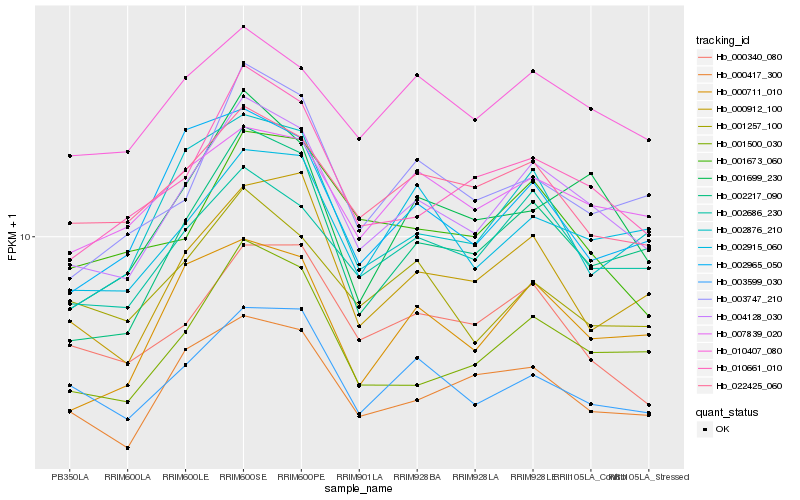
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004128_030 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 8-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002217_090 |
0.0791138615 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 3 |
Hb_010661_010 |
0.0793487596 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 4 |
Hb_002686_230 |
0.082727008 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_001500_030 |
0.0847025798 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 6 |
Hb_010407_080 |
0.0966498623 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003747_210 |
0.0970663024 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 8 |
Hb_003599_030 |
0.0980379468 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000912_100 |
0.0983042394 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 10 |
Hb_001699_230 |
0.0988951026 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 11 |
Hb_002915_060 |
0.0990438031 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1-like [Jatropha curcas] |
| 12 |
Hb_007839_020 |
0.0993213072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_022425_060 |
0.0997396376 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 14 |
Hb_000711_010 |
0.1001393089 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 15 |
Hb_000417_300 |
0.1010115186 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7 [Jatropha curcas] |
| 16 |
Hb_000340_080 |
0.1049636747 |
- |
- |
PREDICTED: isoamylase 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002876_210 |
0.1057698663 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 18 |
Hb_001673_060 |
0.1062080289 |
- |
- |
PREDICTED: uncharacterized protein LOC105635789 [Jatropha curcas] |
| 19 |
Hb_001257_100 |
0.1074563332 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 20 |
Hb_002965_050 |
0.1119090432 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |