| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
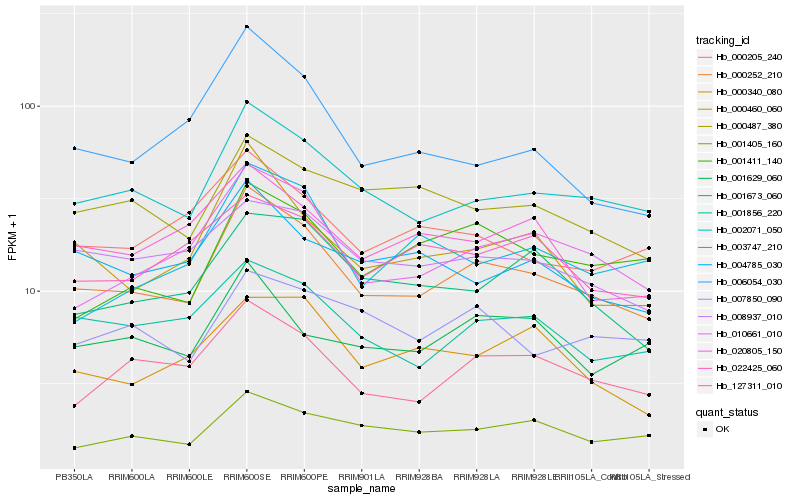
Hb_000252_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644655 [Jatropha curcas] |
| 2 |
Hb_002071_050 |
0.0714052399 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 5-like [Jatropha curcas] |
| 3 |
Hb_010661_010 |
0.0899119938 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 4 |
Hb_127311_010 |
0.0999459409 |
- |
- |
hypothetical protein JCGZ_07497 [Jatropha curcas] |
| 5 |
Hb_000460_060 |
0.1004482821 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 6 |
Hb_001856_220 |
0.1036942296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001405_160 |
0.1122670171 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g09680 [Jatropha curcas] |
| 8 |
Hb_001673_060 |
0.1124569031 |
- |
- |
PREDICTED: uncharacterized protein LOC105635789 [Jatropha curcas] |
| 9 |
Hb_008937_010 |
0.1139645494 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80A2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000487_380 |
0.116094526 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004785_030 |
0.1170124613 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 12 |
Hb_020805_150 |
0.1173479935 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000205_240 |
0.1176962725 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_001629_060 |
0.1204493655 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
Flavin-containing amine oxidase domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_001411_140 |
0.1215639519 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 16 |
Hb_022425_060 |
0.1223435347 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 17 |
Hb_006054_030 |
0.1242275976 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 18 |
Hb_007850_090 |
0.1246166898 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003747_210 |
0.1251305405 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 20 |
Hb_000340_080 |
0.1252308561 |
- |
- |
PREDICTED: isoamylase 2, chloroplastic [Jatropha curcas] |