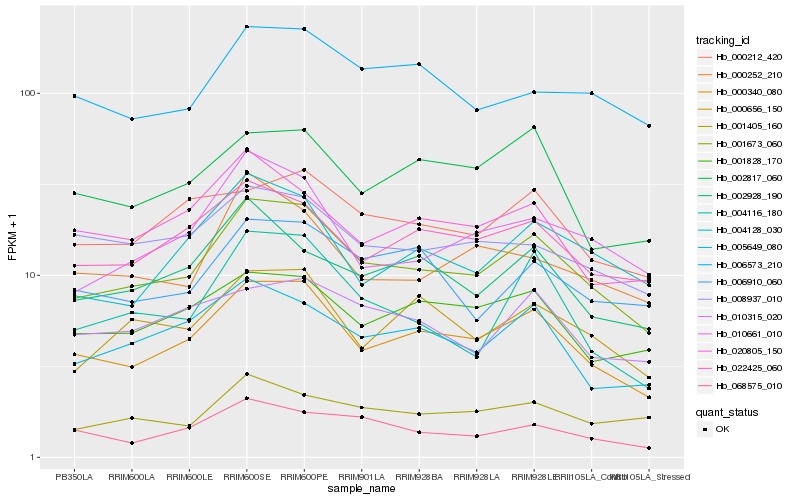
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001673_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635789 [Jatropha curcas] |
| 2 |
Hb_000340_080 |
0.0546698737 |
- |
- |
PREDICTED: isoamylase 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_022425_060 |
0.0967127785 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 4 |
Hb_005649_080 |
0.099465485 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 5 |
Hb_000656_150 |
0.1008467382 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 6 |
Hb_010661_010 |
0.1018581606 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 7 |
Hb_004128_030 |
0.1062080289 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 8-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001828_170 |
0.1064181595 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 9 |
Hb_002928_190 |
0.1066683112 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 10 |
Hb_000212_420 |
0.1072711971 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 11 |
Hb_068575_010 |
0.1076737155 |
- |
- |
PREDICTED: protein GAMETE EXPRESSED 2 [Jatropha curcas] |
| 12 |
Hb_004116_180 |
0.1082347758 |
- |
- |
PREDICTED: uncharacterized protein LOC105646147 [Jatropha curcas] |
| 13 |
Hb_006910_060 |
0.1085246358 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780-like [Jatropha curcas] |
| 14 |
Hb_001405_160 |
0.1088064244 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g09680 [Jatropha curcas] |
| 15 |
Hb_006573_210 |
0.1089796041 |
- |
- |
PREDICTED: protein SGT1 homolog [Jatropha curcas] |
| 16 |
Hb_008937_010 |
0.1121849397 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80A2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000252_210 |
0.1124569031 |
- |
- |
PREDICTED: uncharacterized protein LOC105644655 [Jatropha curcas] |
| 18 |
Hb_010315_020 |
0.1131598734 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 19 |
Hb_020805_150 |
0.113511464 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_002817_060 |
0.1142942836 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |