| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
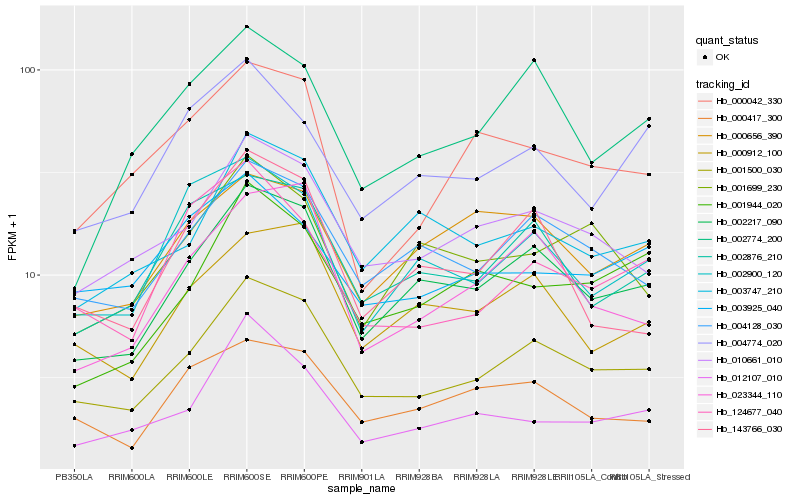
Hb_001500_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002217_090 |
0.0751339543 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 3 |
Hb_010661_010 |
0.0805322742 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 4 |
Hb_004128_030 |
0.0847025798 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 8-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000912_100 |
0.1068409216 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 6 |
Hb_002876_210 |
0.1089031919 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 7 |
Hb_003747_210 |
0.1103927334 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 8 |
Hb_023344_110 |
0.1116827888 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 9 |
Hb_000417_300 |
0.114781538 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7 [Jatropha curcas] |
| 10 |
Hb_001944_020 |
0.1216879924 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 11 |
Hb_124677_040 |
0.1226767669 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 12 |
Hb_012107_010 |
0.1227487473 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 13 |
Hb_003925_040 |
0.1284328206 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 14 |
Hb_000042_330 |
0.1287081462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002900_120 |
0.1301536897 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 16 |
Hb_143766_030 |
0.1304396797 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_001699_230 |
0.1307180167 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 18 |
Hb_002774_200 |
0.1308449399 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 19 |
Hb_000656_390 |
0.1317742162 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004774_020 |
0.1334314686 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |