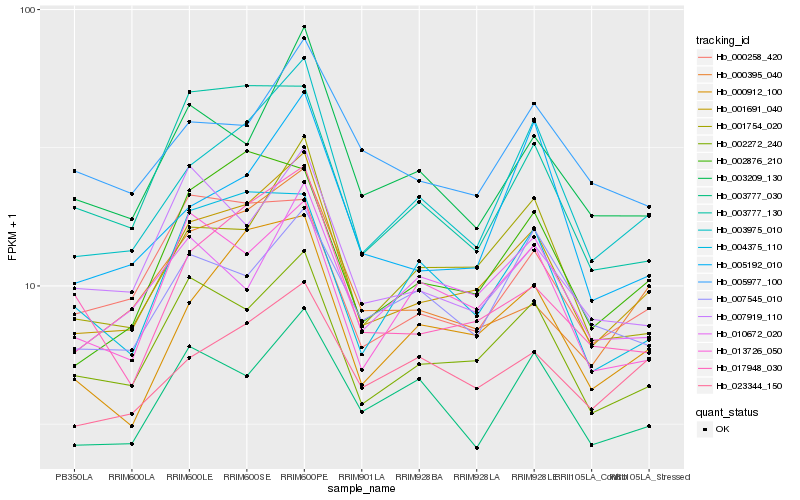
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001691_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649546 isoform X2 [Jatropha curcas] |
| 2 |
Hb_003975_010 |
0.066843507 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 3 |
Hb_002272_240 |
0.0728050693 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_000912_100 |
0.081253803 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 5 |
Hb_001754_020 |
0.0818545272 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 6 |
Hb_000395_040 |
0.0910572666 |
- |
- |
PREDICTED: uncharacterized protein LOC105645576 [Jatropha curcas] |
| 7 |
Hb_002876_210 |
0.0935805048 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 8 |
Hb_003209_130 |
0.1000577574 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 9 |
Hb_017948_030 |
0.100229169 |
- |
- |
PREDICTED: uncharacterized protein LOC105639493 [Jatropha curcas] |
| 10 |
Hb_023344_150 |
0.1007436535 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 11 |
Hb_013726_050 |
0.1016607427 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 12 |
Hb_000258_420 |
0.1027271666 |
- |
- |
sucrose transporter 2A [Hevea brasiliensis] |
| 13 |
Hb_003777_030 |
0.1060361103 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 14 |
Hb_004375_110 |
0.1070604628 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 15 |
Hb_005192_010 |
0.1093899307 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 16 |
Hb_007919_110 |
0.110953277 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 17 |
Hb_005977_100 |
0.1120351725 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 18 |
Hb_003777_130 |
0.112465555 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 19 |
Hb_010672_020 |
0.1134712704 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_007545_010 |
0.1138011832 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |