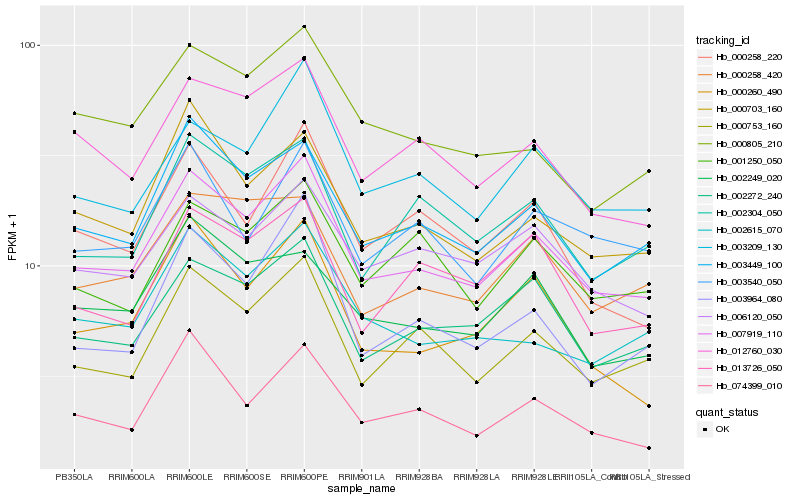
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007919_110 |
0.0 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 2 |
Hb_003449_100 |
0.0684251104 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 3 |
Hb_000753_160 |
0.0722925822 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 4 |
Hb_002272_240 |
0.074790795 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_012760_030 |
0.0798504056 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 10 [Gossypium raimondii] |
| 6 |
Hb_003209_130 |
0.0805375194 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 7 |
Hb_006120_050 |
0.0821044071 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 8 |
Hb_000703_160 |
0.0831564562 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_074399_010 |
0.0853980046 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 10 |
Hb_002304_050 |
0.0869361975 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 11 |
Hb_002615_070 |
0.0872532758 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000805_210 |
0.0911100908 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 13 |
Hb_000258_420 |
0.0920640073 |
- |
- |
sucrose transporter 2A [Hevea brasiliensis] |
| 14 |
Hb_000258_220 |
0.093049173 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000260_490 |
0.0931038655 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 16 |
Hb_001250_050 |
0.0939165116 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003540_050 |
0.0942148844 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 18 |
Hb_013726_050 |
0.0963895235 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 19 |
Hb_003964_080 |
0.0971608696 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 20 |
Hb_002249_020 |
0.0971832455 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |