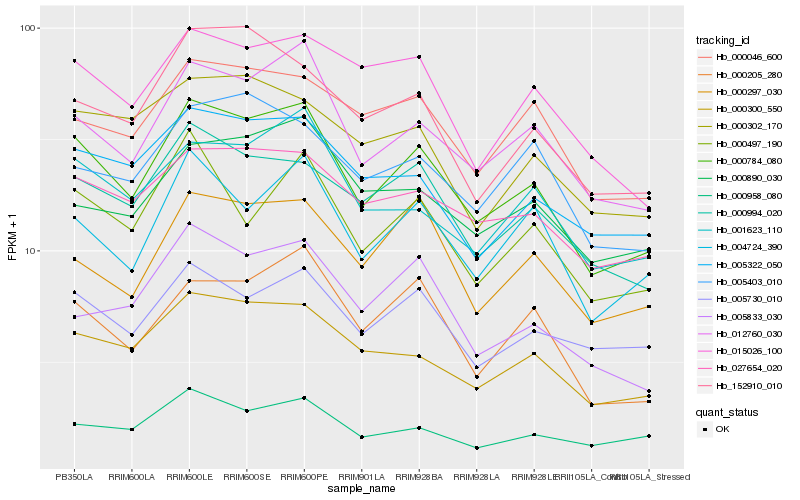
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000784_080 |
0.0 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 2 |
Hb_012760_030 |
0.0677121518 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 10 [Gossypium raimondii] |
| 3 |
Hb_000300_550 |
0.0810174002 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 4 |
Hb_000994_020 |
0.0851217635 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_001623_110 |
0.0893417073 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_027654_020 |
0.0894653379 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 7 |
Hb_004724_390 |
0.0895760559 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 8 |
Hb_000205_280 |
0.0905121 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 9 |
Hb_005322_050 |
0.0922926871 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 10 |
Hb_005730_010 |
0.0962181242 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 11 |
Hb_152910_010 |
0.097851495 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 12 |
Hb_000046_600 |
0.0980649438 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 13 |
Hb_000297_030 |
0.0983298235 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000497_190 |
0.0995935059 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 15 |
Hb_000958_080 |
0.1001519848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_015026_100 |
0.1004356464 |
- |
- |
PREDICTED: transmembrane protein 184C-like [Jatropha curcas] |
| 17 |
Hb_005833_030 |
0.1023671577 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |
| 18 |
Hb_000890_030 |
0.1027463723 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 19 |
Hb_005403_010 |
0.1042496574 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 20 |
Hb_000302_170 |
0.1047287427 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |