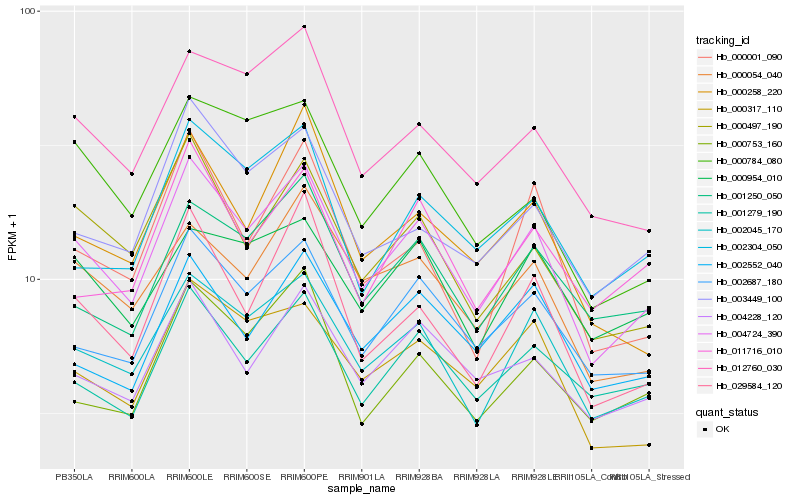
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004724_390 |
0.0 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 2 |
Hb_002045_170 |
0.0573041373 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 3 |
Hb_002687_180 |
0.0708536165 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 4 |
Hb_000497_190 |
0.0800017968 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 5 |
Hb_000317_110 |
0.0807152744 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B1, mitochondrial isoform X2 [Jatropha curcas] |
| 6 |
Hb_004228_120 |
0.0813723465 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 7 |
Hb_012760_030 |
0.0832563221 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 10 [Gossypium raimondii] |
| 8 |
Hb_029584_120 |
0.0846309557 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000054_040 |
0.084991898 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 10 |
Hb_003449_100 |
0.0871806962 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 11 |
Hb_001279_190 |
0.0881292783 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001250_050 |
0.0886361699 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000784_080 |
0.0895760559 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 14 |
Hb_000954_010 |
0.0933757345 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 15 |
Hb_002304_050 |
0.0933809016 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 16 |
Hb_000753_160 |
0.0943069854 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 17 |
Hb_002552_040 |
0.0950036848 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 18 |
Hb_000258_220 |
0.0956009791 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000001_090 |
0.0956057555 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 20 |
Hb_011716_010 |
0.0980239951 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |