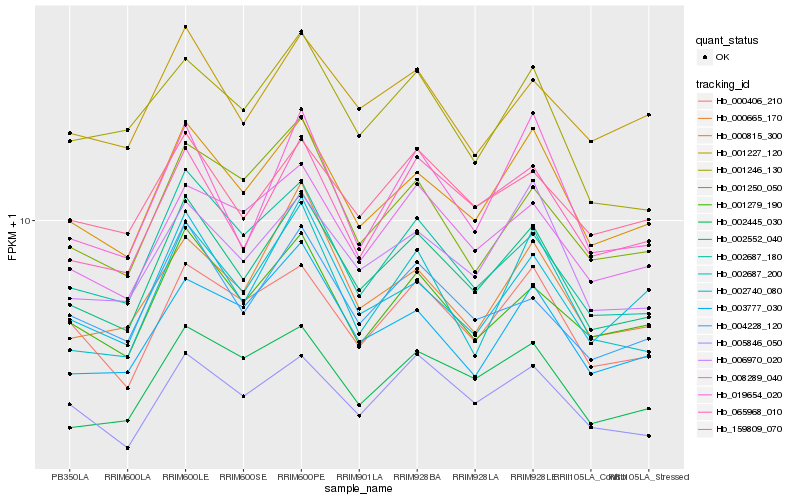
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002552_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 2 |
Hb_000815_300 |
0.0501906924 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 3 |
Hb_002687_180 |
0.0621446083 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 4 |
Hb_004228_120 |
0.0668451741 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 5 |
Hb_001279_190 |
0.0670898097 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 6 |
Hb_065968_010 |
0.0673843352 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 7 |
Hb_006970_020 |
0.0691095994 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 8 |
Hb_008289_040 |
0.071025179 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 9 |
Hb_000406_210 |
0.0727236188 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 10 |
Hb_005846_050 |
0.0742500742 |
- |
- |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 11 |
Hb_002445_030 |
0.0763846772 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 12 |
Hb_002687_200 |
0.0778746537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_159809_070 |
0.0781641225 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 14 |
Hb_001250_050 |
0.0784707749 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001227_120 |
0.0786984568 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 16 |
Hb_000665_170 |
0.0792180481 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 17 |
Hb_019654_020 |
0.0792831529 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 18 |
Hb_003777_030 |
0.0792875372 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 19 |
Hb_001246_130 |
0.0794156663 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 20 |
Hb_002740_080 |
0.0797770306 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |