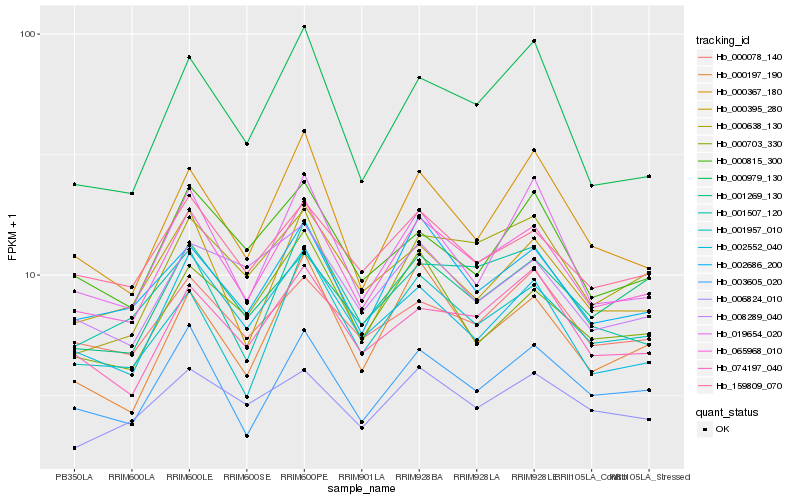
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065968_010 |
0.0 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002686_200 |
0.0605780349 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 3 |
Hb_003605_020 |
0.0641312661 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 4 |
Hb_159809_070 |
0.0653874324 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 5 |
Hb_002552_040 |
0.0673843352 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 6 |
Hb_074197_040 |
0.070152451 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 7 |
Hb_001957_010 |
0.0722565212 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000395_280 |
0.0742276136 |
- |
- |
PREDICTED: thioredoxin-related transmembrane protein 2 [Vitis vinifera] |
| 9 |
Hb_000367_180 |
0.0758111625 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 10 |
Hb_000197_190 |
0.0764982675 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_001269_130 |
0.0772319322 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 12 |
Hb_000078_140 |
0.077862596 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 13 |
Hb_000703_330 |
0.0784963486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_019654_020 |
0.0792840659 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 15 |
Hb_000979_130 |
0.0793969143 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 16 |
Hb_001507_120 |
0.0796895783 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_000815_300 |
0.0802595499 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 18 |
Hb_008289_040 |
0.081220104 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 19 |
Hb_006824_010 |
0.0818120359 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 20 |
Hb_000638_130 |
0.0821280813 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |