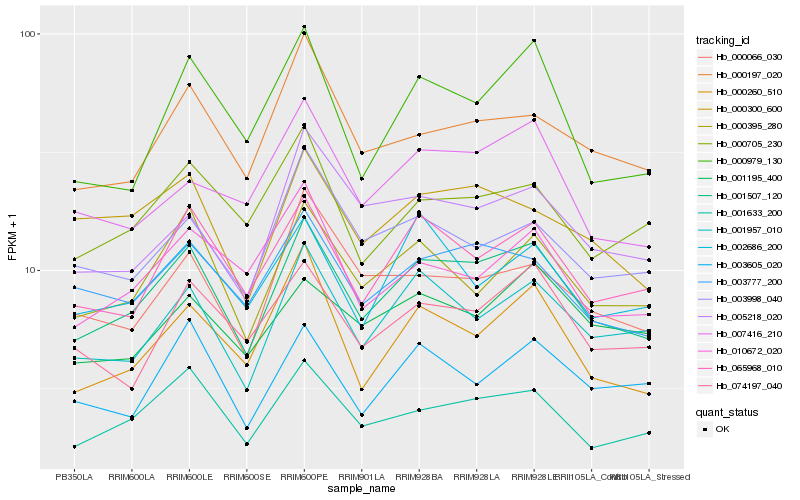
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001507_120 |
0.0 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_001633_200 |
0.0713168247 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |
| 3 |
Hb_001957_010 |
0.0796758248 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 4 |
Hb_065968_010 |
0.0796895783 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000395_280 |
0.0831798047 |
- |
- |
PREDICTED: thioredoxin-related transmembrane protein 2 [Vitis vinifera] |
| 6 |
Hb_003777_200 |
0.085437425 |
- |
- |
PREDICTED: uncharacterized protein LOC105640933 [Jatropha curcas] |
| 7 |
Hb_000260_510 |
0.0911374036 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 8 |
Hb_074197_040 |
0.0929387135 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 9 |
Hb_000197_020 |
0.0947015699 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 10 |
Hb_000979_130 |
0.0949023885 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 11 |
Hb_000705_230 |
0.0971894926 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 12 |
Hb_007416_210 |
0.0972008248 |
- |
- |
PREDICTED: coatomer subunit beta-1 [Jatropha curcas] |
| 13 |
Hb_000300_600 |
0.0975914496 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 14 |
Hb_003605_020 |
0.0993067714 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 15 |
Hb_005218_020 |
0.1003063065 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 16 |
Hb_000066_030 |
0.1010179023 |
- |
- |
PREDICTED: uncharacterized protein LOC105643898 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001195_400 |
0.1014187086 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 18 |
Hb_002686_200 |
0.1016466302 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 19 |
Hb_003998_040 |
0.1020087973 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 20 |
Hb_010672_020 |
0.1020188647 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |