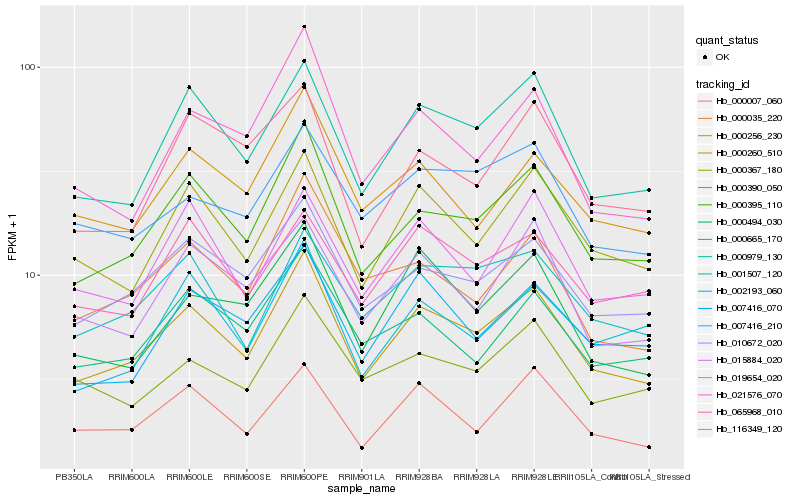
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_510 |
0.0 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 2 |
Hb_000395_110 |
0.0505245938 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 3 |
Hb_000979_130 |
0.0665524899 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 4 |
Hb_010672_020 |
0.0753861008 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_000367_180 |
0.0780548651 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 6 |
Hb_000007_060 |
0.0889254151 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001507_120 |
0.0911374036 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_021576_070 |
0.0919857872 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 9 |
Hb_015884_020 |
0.0925468122 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 10 |
Hb_007416_070 |
0.0925561894 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 11 |
Hb_000494_030 |
0.0931927793 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 12 |
Hb_000665_170 |
0.0935617549 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 13 |
Hb_000256_230 |
0.0935747156 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 14 |
Hb_019654_020 |
0.0959212924 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 15 |
Hb_000035_220 |
0.0961902066 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 16 |
Hb_000390_050 |
0.0963014343 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 17 |
Hb_007416_210 |
0.0971933335 |
- |
- |
PREDICTED: coatomer subunit beta-1 [Jatropha curcas] |
| 18 |
Hb_002193_060 |
0.0992764091 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 19 |
Hb_116349_120 |
0.1003870859 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_065968_010 |
0.1015195582 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |