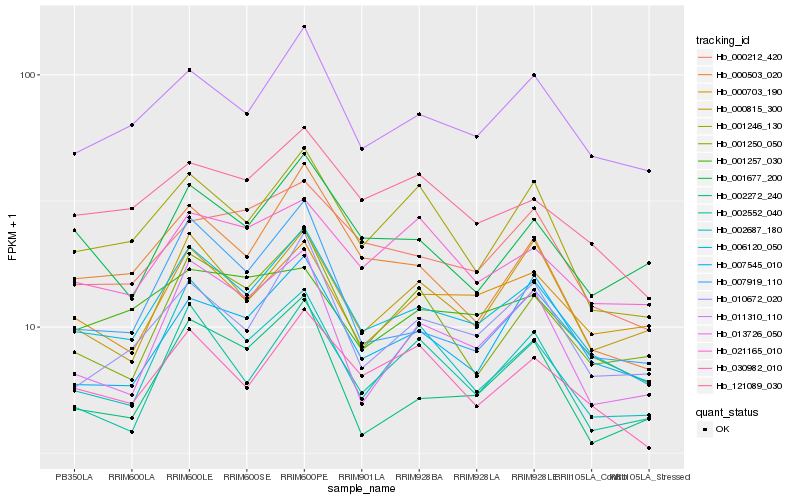
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006120_050 |
0.0 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 2 |
Hb_011310_110 |
0.0545142472 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 3 |
Hb_030982_010 |
0.0713388088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010672_020 |
0.0745467602 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_002272_240 |
0.0750308364 |
- |
- |
catalytic, putative [Ricinus communis] |
| 6 |
Hb_002687_180 |
0.0753509915 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 7 |
Hb_121089_030 |
0.076818173 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 8 |
Hb_001246_130 |
0.0769377011 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000212_420 |
0.0792506098 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 10 |
Hb_002552_040 |
0.0802742382 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 11 |
Hb_000503_020 |
0.0802977334 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 12 |
Hb_000703_190 |
0.0809463794 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001250_050 |
0.0819530989 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007919_110 |
0.0821044071 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 15 |
Hb_000815_300 |
0.0823719113 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 16 |
Hb_007545_010 |
0.083777757 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 17 |
Hb_013726_050 |
0.0838735724 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 18 |
Hb_021165_010 |
0.0848213415 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001257_030 |
0.0862492983 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek5 [Jatropha curcas] |
| 20 |
Hb_001677_200 |
0.0872169102 |
- |
- |
conserved hypothetical protein [Ricinus communis] |