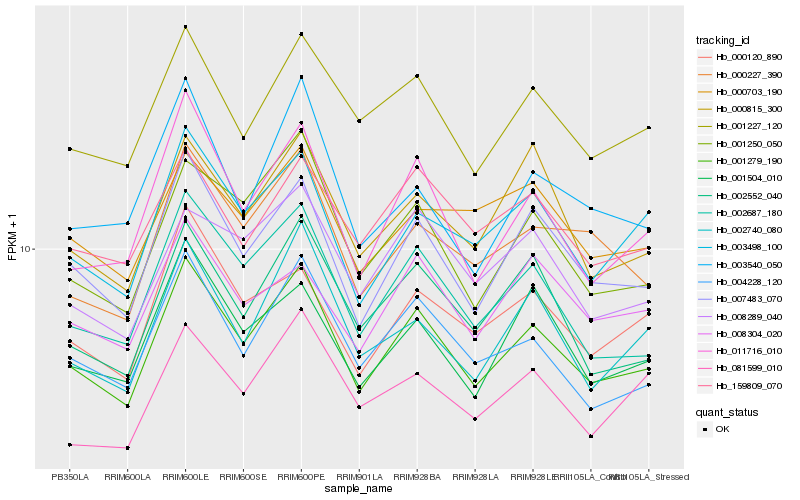
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001279_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007483_070 |
0.0449988985 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 3 |
Hb_002687_180 |
0.0552807829 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 4 |
Hb_004228_120 |
0.05840575 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 5 |
Hb_011716_010 |
0.0595710435 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 6 |
Hb_001250_050 |
0.062916398 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001227_120 |
0.0629378981 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 8 |
Hb_002552_040 |
0.0670898097 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 9 |
Hb_003498_100 |
0.067838744 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 10 |
Hb_001504_010 |
0.0692847692 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 11 |
Hb_000227_390 |
0.0716168196 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002740_080 |
0.0723593073 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 13 |
Hb_000815_300 |
0.072600586 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 14 |
Hb_000120_890 |
0.0742804271 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 15 |
Hb_008289_040 |
0.0753376736 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 16 |
Hb_159809_070 |
0.0759111689 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 17 |
Hb_008304_020 |
0.0805094278 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 18 |
Hb_003540_050 |
0.0809518731 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 19 |
Hb_081599_010 |
0.082125826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000703_190 |
0.0821416117 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |