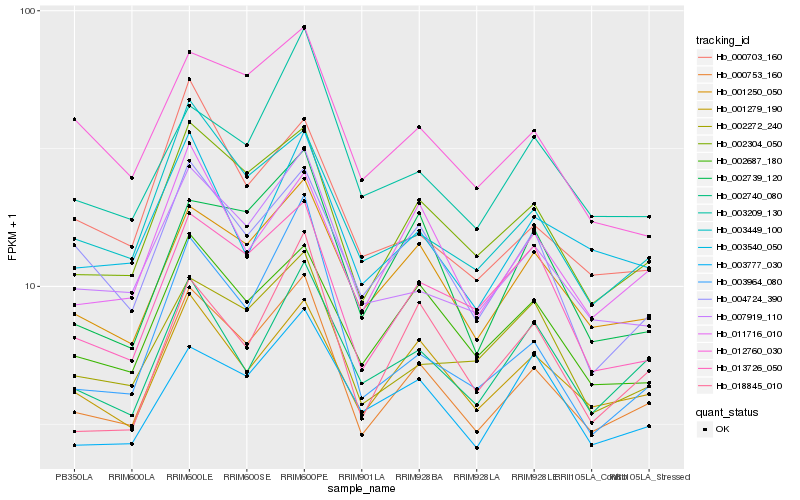
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000753_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 2 |
Hb_002304_050 |
0.0510283292 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 3 |
Hb_001250_050 |
0.0682426932 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007919_110 |
0.0722925822 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 5 |
Hb_003449_100 |
0.0762196951 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 6 |
Hb_002687_180 |
0.0812775014 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 7 |
Hb_001279_190 |
0.0855881611 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 8 |
Hb_011716_010 |
0.0860321281 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 9 |
Hb_013726_050 |
0.0878593315 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 10 |
Hb_003964_080 |
0.0904822054 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 11 |
Hb_002740_080 |
0.0922586183 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 12 |
Hb_002272_240 |
0.0925085189 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_002739_120 |
0.0928126856 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 14 |
Hb_004724_390 |
0.0943069854 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 15 |
Hb_003540_050 |
0.0946167312 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 16 |
Hb_003209_130 |
0.0948960012 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 17 |
Hb_018845_010 |
0.0965843044 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 18 |
Hb_000703_160 |
0.0986180689 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003777_030 |
0.0987897006 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 20 |
Hb_012760_030 |
0.0989594931 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 10 [Gossypium raimondii] |