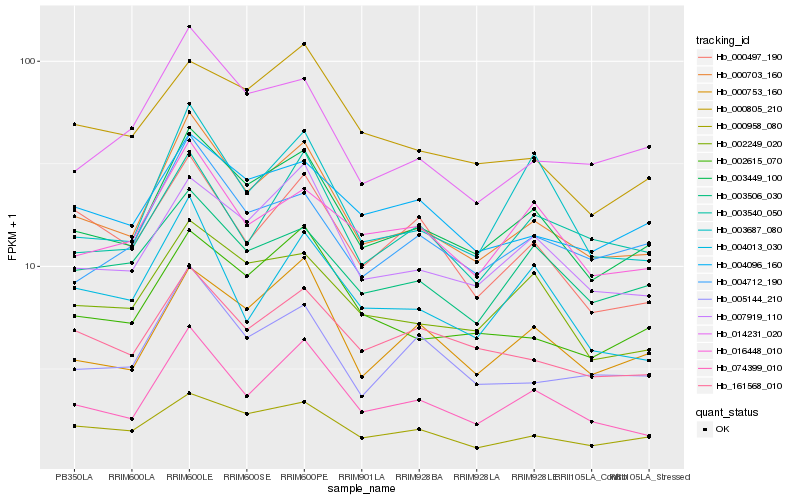
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_160 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_003449_100 |
0.0519311196 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 3 |
Hb_074399_010 |
0.0674305697 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 4 |
Hb_007919_110 |
0.0831564562 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 5 |
Hb_005144_210 |
0.0869585578 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_161568_010 |
0.0873834551 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 7 |
Hb_000958_080 |
0.087481541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002615_070 |
0.0877590172 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 9 |
Hb_014231_020 |
0.0921324319 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 10 |
Hb_002249_020 |
0.0963193574 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 11 |
Hb_004712_190 |
0.0974499434 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 12 |
Hb_004096_160 |
0.0984018895 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 13 |
Hb_000753_160 |
0.0986180689 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 14 |
Hb_003506_030 |
0.0996198215 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 15 |
Hb_016448_010 |
0.1017263277 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 16 |
Hb_000497_190 |
0.1020939482 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 17 |
Hb_003540_050 |
0.1030590923 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 18 |
Hb_003687_080 |
0.1054867931 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 19 |
Hb_004013_030 |
0.1075931621 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 20 |
Hb_000805_210 |
0.1084019492 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |