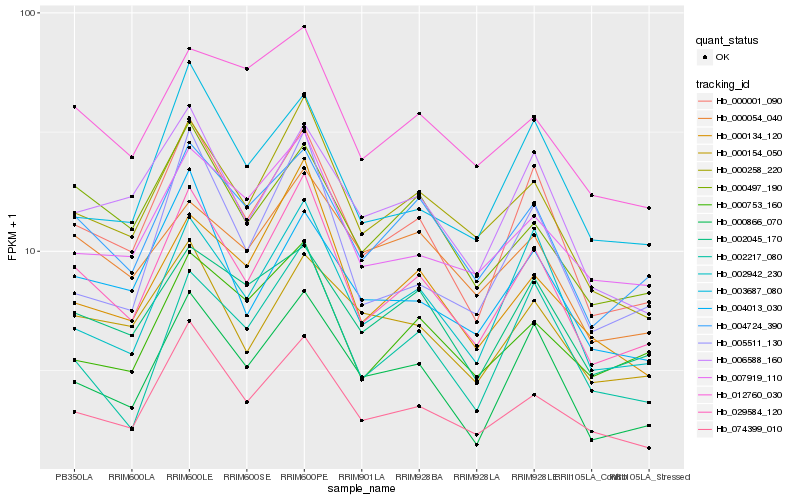
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029584_120 |
0.0 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000001_090 |
0.0624192547 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 3 |
Hb_000258_220 |
0.0701744142 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_074399_010 |
0.080997335 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 5 |
Hb_004724_390 |
0.0846309557 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 6 |
Hb_000866_070 |
0.0871457741 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 7 |
Hb_002942_230 |
0.0934950786 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 8 |
Hb_000134_120 |
0.0966292345 |
- |
- |
PREDICTED: uncharacterized protein LOC105628669 [Jatropha curcas] |
| 9 |
Hb_000497_190 |
0.1004075226 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 10 |
Hb_007919_110 |
0.1004807723 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 11 |
Hb_005511_130 |
0.1036446129 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 12 |
Hb_000753_160 |
0.1062775795 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 13 |
Hb_006588_160 |
0.1071568003 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_002045_170 |
0.108379027 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 15 |
Hb_004013_030 |
0.1100439584 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 16 |
Hb_003687_080 |
0.1104225296 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 17 |
Hb_002217_080 |
0.1104460882 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 16 [Jatropha curcas] |
| 18 |
Hb_012760_030 |
0.1114295828 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 10 [Gossypium raimondii] |
| 19 |
Hb_000154_050 |
0.1128988928 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 20 |
Hb_000054_040 |
0.1132035974 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |