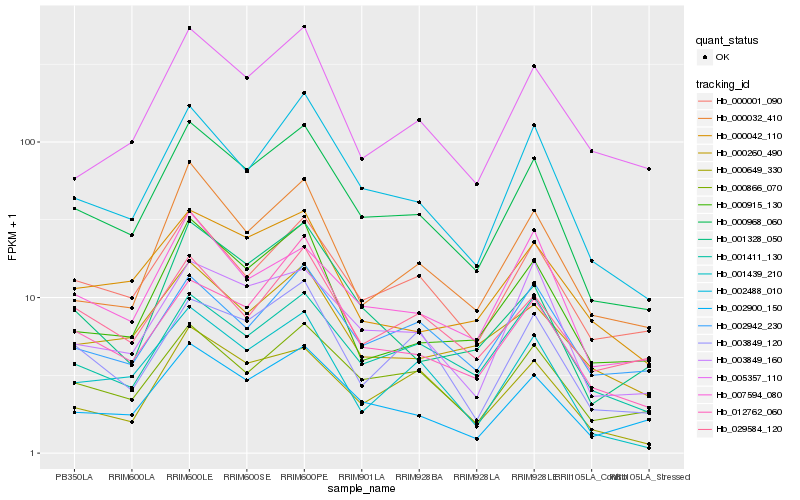
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000968_060 |
0.0 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 2 |
Hb_002488_010 |
0.0754659984 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 3 |
Hb_002900_150 |
0.0881828228 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 4 |
Hb_000866_070 |
0.0952674748 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 5 |
Hb_000001_090 |
0.0991739813 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 6 |
Hb_003849_160 |
0.0997035397 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 7 |
Hb_000032_410 |
0.1068368909 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 8 |
Hb_001411_130 |
0.1103841962 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000915_130 |
0.1117523231 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 10 |
Hb_007594_080 |
0.1118215695 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 11 |
Hb_000260_490 |
0.1126003283 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 12 |
Hb_003849_120 |
0.1136460175 |
- |
- |
PREDICTED: uncharacterized protein LOC100264440 [Vitis vinifera] |
| 13 |
Hb_001439_210 |
0.1140109219 |
- |
- |
EXS family protein [Populus trichocarpa] |
| 14 |
Hb_000649_330 |
0.1144649401 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 15 |
Hb_000042_110 |
0.1161105968 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 16 |
Hb_001328_050 |
0.1169118381 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 17 |
Hb_012762_060 |
0.1172776741 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 18 |
Hb_029584_120 |
0.1211012724 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002942_230 |
0.1218070607 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 20 |
Hb_005357_110 |
0.121848573 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |