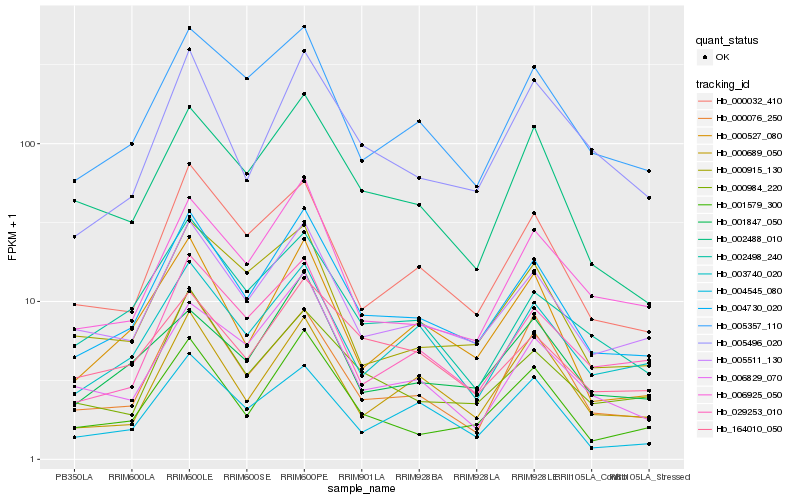
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004730_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 2 |
Hb_005511_130 |
0.0693290521 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 3 |
Hb_001579_300 |
0.0882930043 |
- |
- |
PREDICTED: probable polygalacturonase At1g80170 [Jatropha curcas] |
| 4 |
Hb_000032_410 |
0.0927439141 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 5 |
Hb_005357_110 |
0.0929679511 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 6 |
Hb_000527_080 |
0.0994086032 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 7 |
Hb_003740_020 |
0.1006233479 |
- |
- |
PREDICTED: methylglutaconyl-CoA hydratase, mitochondrial isoform X2 [Jatropha curcas] |
| 8 |
Hb_000984_220 |
0.1009333026 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 9 |
Hb_006925_050 |
0.1020723311 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 10 |
Hb_029253_010 |
0.1033834176 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 11 |
Hb_002488_010 |
0.1093587085 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 12 |
Hb_005496_020 |
0.1114281455 |
- |
- |
PREDICTED: uncharacterized protein LOC105630867 [Jatropha curcas] |
| 13 |
Hb_000915_130 |
0.1117492705 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 14 |
Hb_002498_240 |
0.1122034323 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001847_050 |
0.1149347013 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |
| 16 |
Hb_164010_050 |
0.1169579608 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 17 |
Hb_004545_080 |
0.1183157266 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_006829_070 |
0.1191996144 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 19 |
Hb_000076_250 |
0.119431079 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000689_050 |
0.1201107431 |
- |
- |
conserved hypothetical protein [Ricinus communis] |