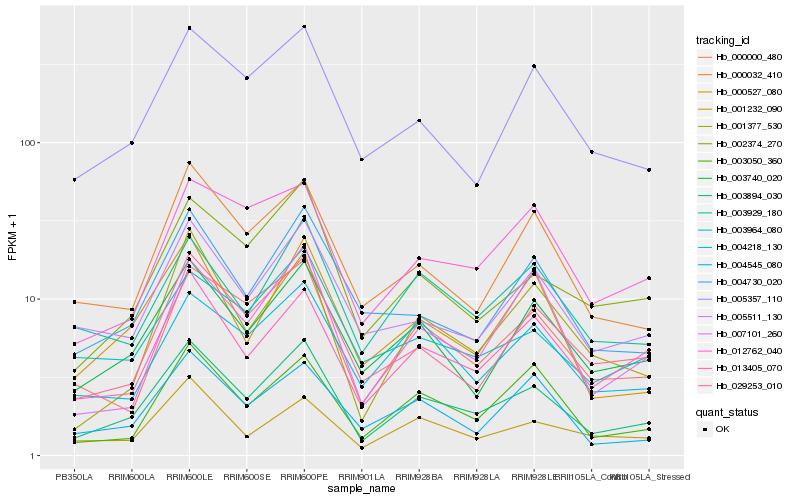
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003894_030 |
0.0 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 2 |
Hb_003740_020 |
0.1081807139 |
- |
- |
PREDICTED: methylglutaconyl-CoA hydratase, mitochondrial isoform X2 [Jatropha curcas] |
| 3 |
Hb_013405_070 |
0.1106210863 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 4 |
Hb_002374_270 |
0.1111661648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_029253_010 |
0.1121300971 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 6 |
Hb_000000_480 |
0.1191981789 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000527_080 |
0.1196292397 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 8 |
Hb_005511_130 |
0.119658888 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 9 |
Hb_000032_410 |
0.1220938867 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 10 |
Hb_001377_530 |
0.1225251853 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 11 |
Hb_005357_110 |
0.1229881391 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 12 |
Hb_004730_020 |
0.1236966452 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 13 |
Hb_003050_360 |
0.1247857485 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_004218_130 |
0.1273644262 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 15 |
Hb_003929_180 |
0.1275242584 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 16 |
Hb_001232_090 |
0.1296053863 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 17 |
Hb_004545_080 |
0.1332330973 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_003964_080 |
0.1347324541 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 19 |
Hb_007101_260 |
0.1372224523 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 20 |
Hb_012762_040 |
0.1378489238 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |