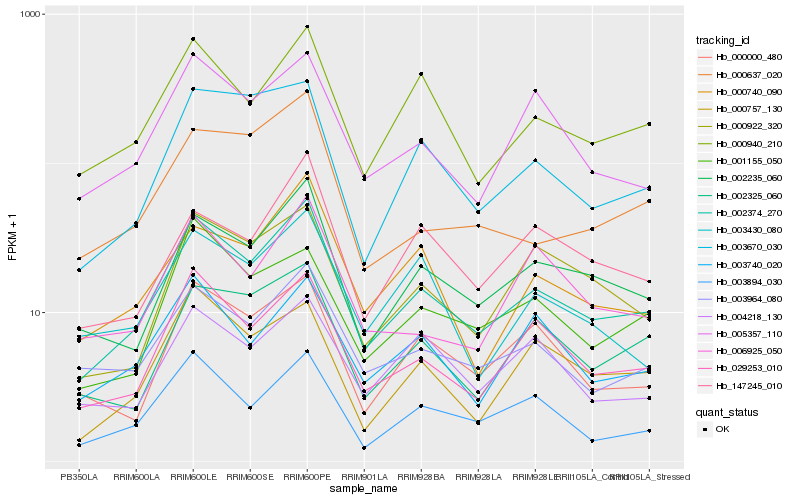
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_270 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_029253_010 |
0.0954439408 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 3 |
Hb_002235_060 |
0.0988540803 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 4 |
Hb_000940_210 |
0.1084636954 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 5 |
Hb_003964_080 |
0.1092976657 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 6 |
Hb_003894_030 |
0.1111661648 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 7 |
Hb_005357_110 |
0.1219818301 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 8 |
Hb_003670_030 |
0.1222215361 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 9 |
Hb_000000_480 |
0.1234172106 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002325_060 |
0.1256788492 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 11 |
Hb_000757_130 |
0.127363604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003740_020 |
0.1274514453 |
- |
- |
PREDICTED: methylglutaconyl-CoA hydratase, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_000922_320 |
0.1284393019 |
- |
- |
hypothetical protein B456_001G071400 [Gossypium raimondii] |
| 14 |
Hb_000740_090 |
0.1299055838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001155_050 |
0.1347303779 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 16 |
Hb_006925_050 |
0.138862596 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 17 |
Hb_147245_010 |
0.1390758982 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 18 |
Hb_000637_020 |
0.1396681939 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 19 |
Hb_004218_130 |
0.139922747 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 20 |
Hb_003430_080 |
0.1421964342 |
- |
- |
zinc finger protein, putative [Ricinus communis] |