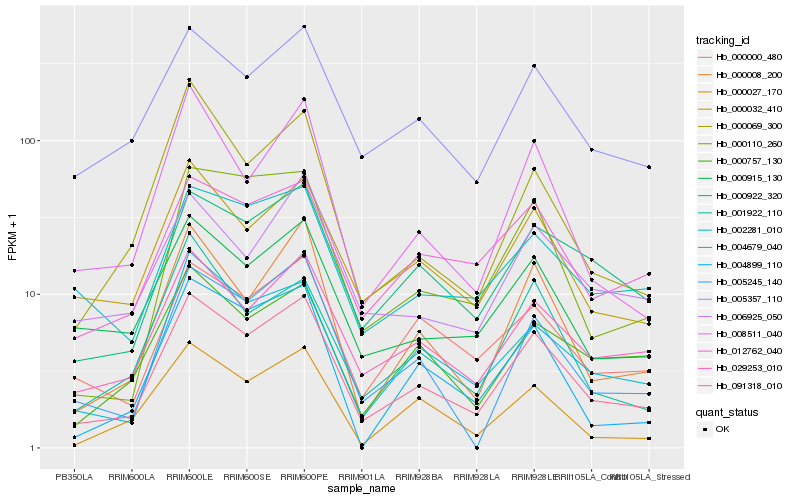
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_091318_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |
| 2 |
Hb_029253_010 |
0.0942987623 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 3 |
Hb_000008_200 |
0.1039461735 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 4 |
Hb_004679_040 |
0.1050623483 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 5 |
Hb_005357_110 |
0.1065079109 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 6 |
Hb_000032_410 |
0.1069270091 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 7 |
Hb_000915_130 |
0.1138284209 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 8 |
Hb_000922_320 |
0.1172653975 |
- |
- |
hypothetical protein B456_001G071400 [Gossypium raimondii] |
| 9 |
Hb_008511_040 |
0.1189471493 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 10 |
Hb_000000_480 |
0.1191352886 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005245_140 |
0.1225701349 |
- |
- |
PREDICTED: ribonuclease 2 [Jatropha curcas] |
| 12 |
Hb_000757_130 |
0.1280457507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000110_260 |
0.1302997444 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 14 |
Hb_012762_040 |
0.1304935252 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 15 |
Hb_000069_300 |
0.1320304807 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 16 |
Hb_001922_110 |
0.1338633467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002281_010 |
0.1348197195 |
- |
- |
hypothetical protein POPTR_0001s47550g [Populus trichocarpa] |
| 18 |
Hb_000027_170 |
0.1350646199 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 19 |
Hb_006925_050 |
0.1352060177 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 20 |
Hb_004899_110 |
0.1359741722 |
- |
- |
PREDICTED: uncharacterized protein LOC105638015 [Jatropha curcas] |