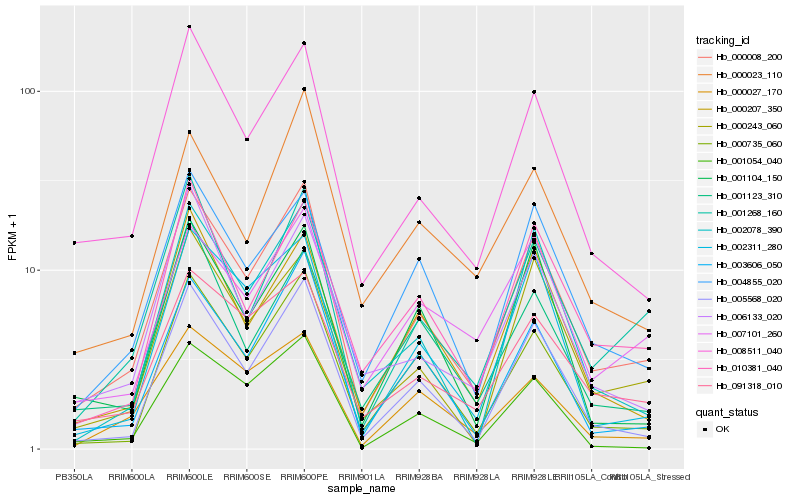
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_200 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 2 |
Hb_005568_020 |
0.0882230624 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008511_040 |
0.0946235801 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 4 |
Hb_003606_050 |
0.102591335 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |
| 5 |
Hb_091318_010 |
0.1039461735 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |
| 6 |
Hb_001268_160 |
0.1105225757 |
- |
- |
Glycerophosphodiester phosphodiesterase [Medicago truncatula] |
| 7 |
Hb_000735_060 |
0.1116734127 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 8 |
Hb_000243_060 |
0.1152127845 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 9 |
Hb_004855_020 |
0.1162154096 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 10 |
Hb_010381_040 |
0.1182919369 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 11 |
Hb_002311_280 |
0.119135117 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 12 |
Hb_000207_350 |
0.1195467998 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 13 |
Hb_002078_390 |
0.1220254927 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 14 |
Hb_000027_170 |
0.1281991744 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 15 |
Hb_001104_150 |
0.1312146825 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 16 |
Hb_001054_040 |
0.131377824 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 17 |
Hb_006133_020 |
0.1337895896 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 18 |
Hb_007101_260 |
0.1340317211 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 19 |
Hb_001123_310 |
0.1350119487 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 20 |
Hb_000023_110 |
0.1365278564 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |