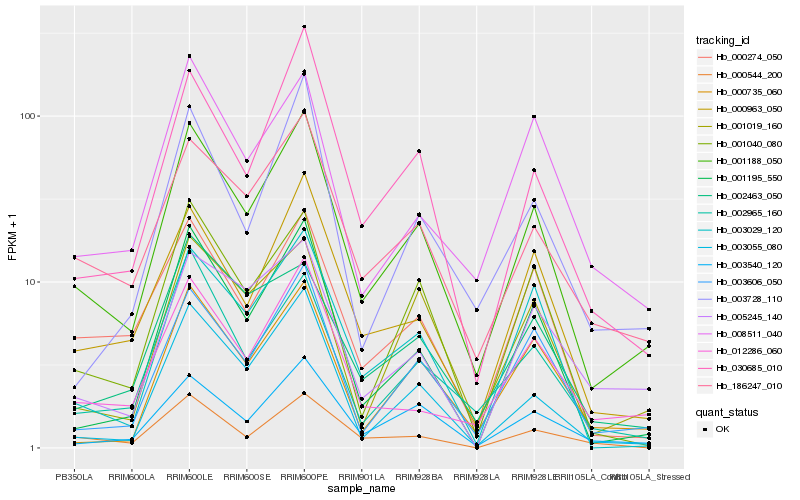
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001188_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_186247_010 |
0.1058888632 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |
| 3 |
Hb_003606_050 |
0.1078086224 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |
| 4 |
Hb_000963_050 |
0.1184537617 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |
| 5 |
Hb_003540_120 |
0.1186244906 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 6 |
Hb_030685_010 |
0.1239924757 |
- |
- |
- |
| 7 |
Hb_000274_050 |
0.1258821212 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 8 |
Hb_001195_550 |
0.1266744926 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 9 |
Hb_012286_060 |
0.1295985044 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_001040_080 |
0.1298296687 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 11 |
Hb_003029_120 |
0.1300001159 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 12 |
Hb_003055_080 |
0.1319654952 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002965_160 |
0.1336803033 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 14 |
Hb_000735_060 |
0.1344757185 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 15 |
Hb_001019_160 |
0.1413778502 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_008511_040 |
0.1446912077 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 17 |
Hb_003728_110 |
0.1473515399 |
- |
- |
PREDICTED: calmodulin-like protein 3 [Populus euphratica] |
| 18 |
Hb_005245_140 |
0.1478519496 |
- |
- |
PREDICTED: ribonuclease 2 [Jatropha curcas] |
| 19 |
Hb_002463_050 |
0.1488489421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000544_200 |
0.1512403811 |
- |
- |
PREDICTED: uncharacterized protein LOC105647171 [Jatropha curcas] |