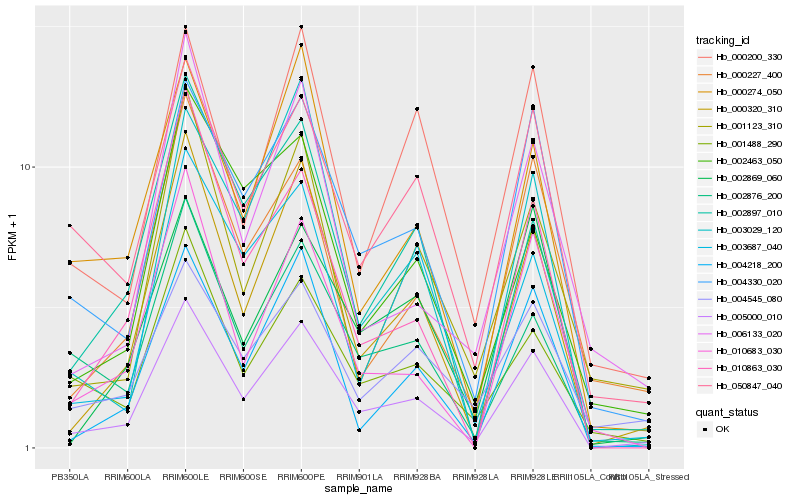
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005000_010 |
0.0 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000320_310 |
0.0848071778 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C [Jatropha curcas] |
| 3 |
Hb_004330_020 |
0.1091592048 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 4 |
Hb_000274_050 |
0.1282494517 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 5 |
Hb_004218_200 |
0.1314337013 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 6 |
Hb_010863_030 |
0.1334749683 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 7 |
Hb_002876_200 |
0.1361821397 |
- |
- |
PREDICTED: uncharacterized protein LOC105641209 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002897_010 |
0.1379886969 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g58300 [Jatropha curcas] |
| 9 |
Hb_050847_040 |
0.138675567 |
- |
- |
PREDICTED: uncharacterized protein LOC105649141 [Jatropha curcas] |
| 10 |
Hb_000227_400 |
0.141535546 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 11 |
Hb_010683_030 |
0.1418402706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002869_060 |
0.14184203 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 13 |
Hb_001488_290 |
0.1435346136 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 14 |
Hb_003029_120 |
0.1473068723 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 15 |
Hb_000200_330 |
0.1473313953 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 16 |
Hb_003687_040 |
0.1474536462 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |
| 17 |
Hb_004545_080 |
0.1478138013 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_002463_050 |
0.1519773902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001123_310 |
0.1523120415 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 20 |
Hb_006133_020 |
0.1534924209 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |