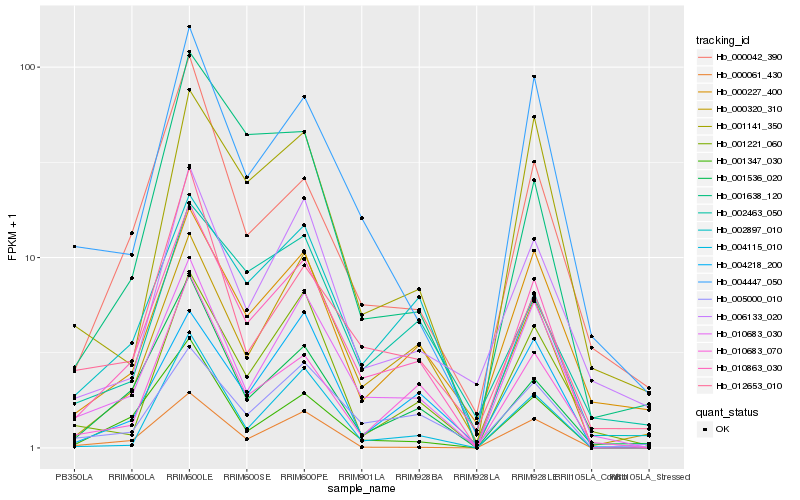
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010863_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 2 |
Hb_010683_030 |
0.1184708662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000320_310 |
0.1288353789 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C [Jatropha curcas] |
| 4 |
Hb_005000_010 |
0.1334749683 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002897_010 |
0.1404689403 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g58300 [Jatropha curcas] |
| 6 |
Hb_010683_070 |
0.1426456602 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001536_020 |
0.1430096571 |
transcription factor |
TF Family: GRF |
PREDICTED: uncharacterized protein LOC105631006 [Jatropha curcas] |
| 8 |
Hb_001347_030 |
0.1444082972 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_000227_400 |
0.1571266502 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 10 |
Hb_004447_050 |
0.1592614624 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004218_200 |
0.1594103231 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 12 |
Hb_004115_010 |
0.1652554378 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 13 |
Hb_001221_060 |
0.1683648349 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 14 |
Hb_000061_430 |
0.1716383126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000042_390 |
0.1717855431 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7 [Jatropha curcas] |
| 16 |
Hb_001638_120 |
0.1719391822 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 17 |
Hb_002463_050 |
0.1725399118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_012653_010 |
0.1729920025 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 19 |
Hb_006133_020 |
0.1745114083 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 20 |
Hb_001141_350 |
0.1749834208 |
- |
- |
hypothetical protein JCGZ_02174 [Jatropha curcas] |