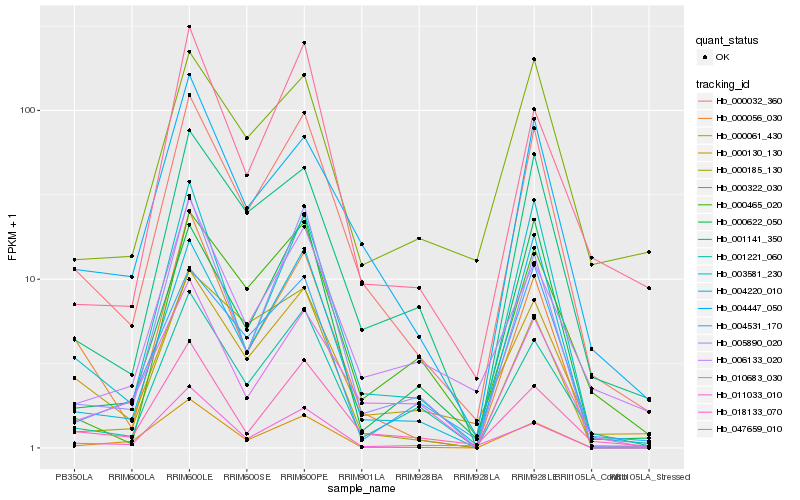
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_360 |
0.0 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit S, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000130_130 |
0.1043623201 |
- |
- |
hypothetical protein POPTR_0001s40330g [Populus trichocarpa] |
| 3 |
Hb_004447_050 |
0.1137273326 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003581_230 |
0.1222488437 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 5 |
Hb_001141_350 |
0.1246472375 |
- |
- |
hypothetical protein JCGZ_02174 [Jatropha curcas] |
| 6 |
Hb_018133_070 |
0.1274660491 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 7 |
Hb_001221_060 |
0.1350292146 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 8 |
Hb_004220_010 |
0.1401823183 |
- |
- |
PREDICTED: uncharacterized protein LOC105642366 [Jatropha curcas] |
| 9 |
Hb_000056_030 |
0.1402011887 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g14390 isoform X1 [Jatropha curcas] |
| 10 |
Hb_010683_030 |
0.1405554243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005890_020 |
0.1442282811 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 12 |
Hb_000622_050 |
0.1469813259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004531_170 |
0.1543543628 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 14 |
Hb_000185_130 |
0.1554683408 |
- |
- |
UBX domain-containing protein 8-B, putative [Ricinus communis] |
| 15 |
Hb_000465_020 |
0.155487497 |
- |
- |
PREDICTED: uncharacterized protein LOC105632280 [Jatropha curcas] |
| 16 |
Hb_006133_020 |
0.156095578 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 17 |
Hb_000061_430 |
0.1567151767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_047659_010 |
0.157395694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000322_030 |
0.1588176688 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 20 |
Hb_011033_010 |
0.1608548234 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |