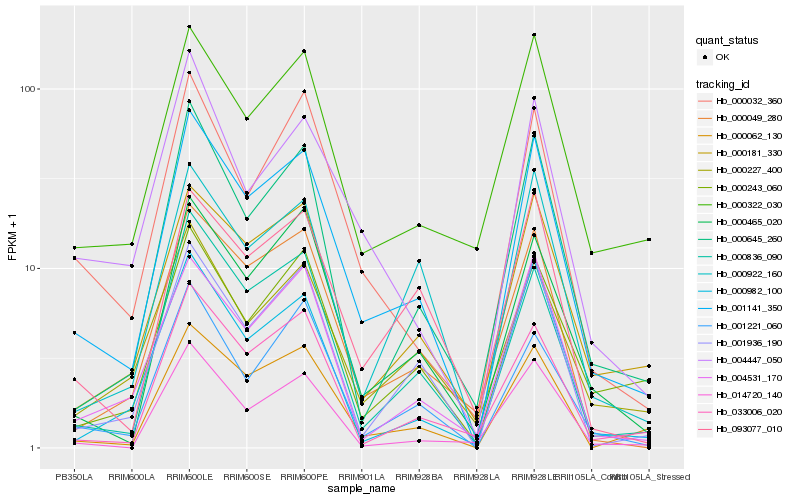
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_350 |
0.0 |
- |
- |
hypothetical protein JCGZ_02174 [Jatropha curcas] |
| 2 |
Hb_000836_090 |
0.1088617565 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 3 |
Hb_000062_130 |
0.1118497249 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_001936_190 |
0.1126097784 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 5 |
Hb_000645_260 |
0.1163548207 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 6 |
Hb_000181_330 |
0.1170605504 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 7 |
Hb_000465_020 |
0.1172221566 |
- |
- |
PREDICTED: uncharacterized protein LOC105632280 [Jatropha curcas] |
| 8 |
Hb_000049_280 |
0.1178483959 |
- |
- |
PREDICTED: ribokinase-like isoform X4 [Jatropha curcas] |
| 9 |
Hb_000032_360 |
0.1246472375 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit S, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004531_170 |
0.1267364641 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 11 |
Hb_000982_100 |
0.1268637801 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PIF1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000322_030 |
0.1270034896 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 13 |
Hb_093077_010 |
0.1288161145 |
transcription factor |
TF Family: G2-like |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_033006_020 |
0.1306028669 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |
| 15 |
Hb_000243_060 |
0.1315377196 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 16 |
Hb_001221_060 |
0.1318227306 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 17 |
Hb_000922_160 |
0.132379383 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 18 |
Hb_000227_400 |
0.1331191707 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 19 |
Hb_004447_050 |
0.1351759703 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 20 |
Hb_014720_140 |
0.1360766008 |
- |
- |
PREDICTED: uncharacterized protein LOC105640367 [Jatropha curcas] |