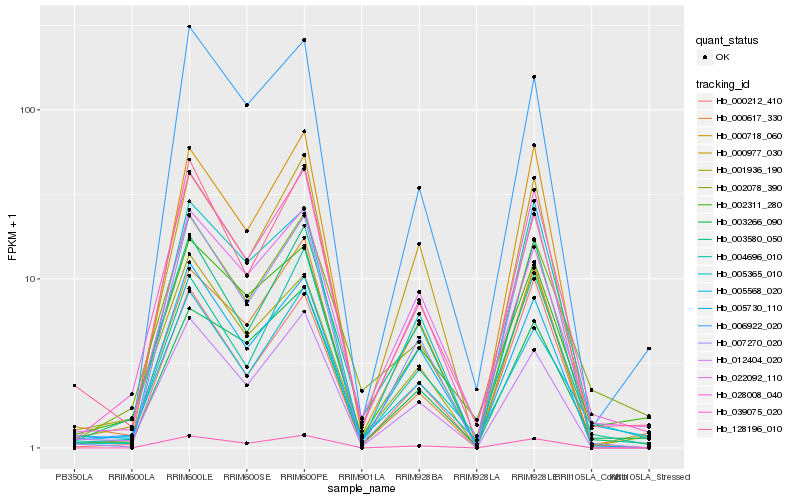
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028008_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003580_050 |
0.0782080371 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 3 |
Hb_002078_390 |
0.0917752992 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 4 |
Hb_004696_010 |
0.0946137374 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 5 |
Hb_001936_190 |
0.0947858754 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 6 |
Hb_012404_020 |
0.0972657614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005365_010 |
0.0973152119 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 8 |
Hb_039075_020 |
0.0989743908 |
- |
- |
PREDICTED: uncharacterized protein LOC103331245 [Prunus mume] |
| 9 |
Hb_003266_090 |
0.1016696559 |
- |
- |
PREDICTED: basic 7S globulin 2-like [Jatropha curcas] |
| 10 |
Hb_005730_110 |
0.1019350293 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 11 |
Hb_002311_280 |
0.1020655421 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 12 |
Hb_005568_020 |
0.105848467 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000718_060 |
0.1060603948 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_007270_020 |
0.1071869048 |
- |
- |
hypothetical protein JCGZ_09593 [Jatropha curcas] |
| 15 |
Hb_128196_010 |
0.1076924631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000212_410 |
0.1099557866 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 17 |
Hb_000977_030 |
0.1111027823 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000617_330 |
0.1112773652 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 19 |
Hb_022092_110 |
0.1124094803 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 20 |
Hb_006922_020 |
0.1124239791 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |