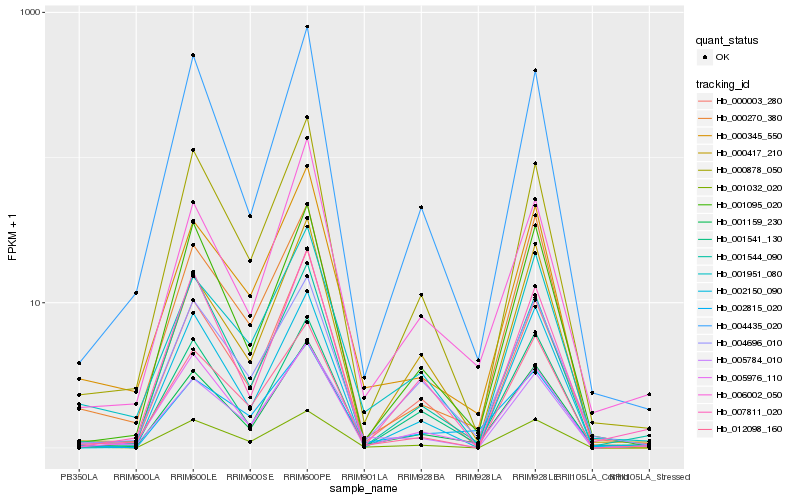
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001159_230 |
0.0 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 2 |
Hb_000878_050 |
0.0926344613 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 3 |
Hb_006002_050 |
0.0930688345 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 4 |
Hb_001095_020 |
0.0948924981 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |
| 5 |
Hb_004696_010 |
0.0964273662 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 6 |
Hb_004435_020 |
0.0991493956 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 7 |
Hb_005976_110 |
0.0991702044 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 8 |
Hb_000417_210 |
0.1009641169 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_000003_280 |
0.1014128009 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001541_130 |
0.1020646884 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 11 |
Hb_001951_080 |
0.1061896718 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 12 |
Hb_005784_010 |
0.1070204098 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 13 |
Hb_002815_020 |
0.1088944785 |
transcription factor |
TF Family: G2-like |
Homeodomain-like superfamily protein isoform 2 [Theobroma cacao] |
| 14 |
Hb_001544_090 |
0.1106650232 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 15 |
Hb_000345_550 |
0.1112320183 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 16 |
Hb_002150_090 |
0.111569613 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 17 |
Hb_007811_020 |
0.111694247 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 18 |
Hb_012098_160 |
0.1126210569 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 19 |
Hb_001032_020 |
0.1127246718 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 20 |
Hb_000270_380 |
0.113160808 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |