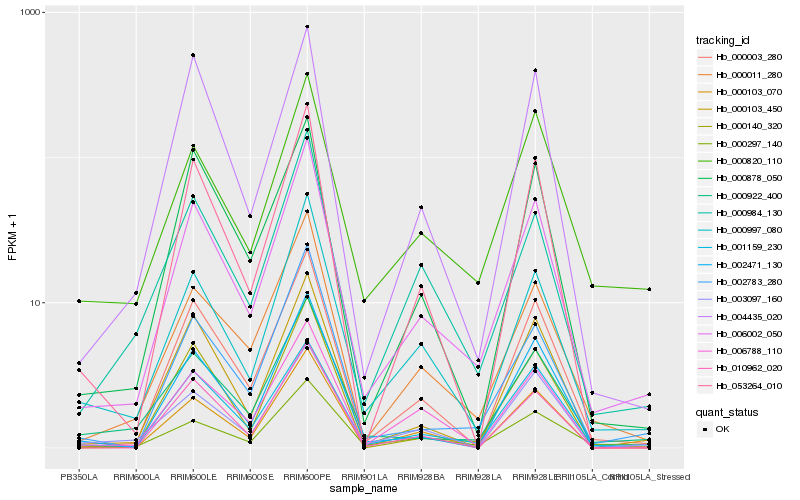
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006002_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 2 |
Hb_000011_280 |
0.0866486882 |
- |
- |
pectinesterase family protein [Populus trichocarpa] |
| 3 |
Hb_000997_080 |
0.0897721903 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001159_230 |
0.0930688345 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 5 |
Hb_000003_280 |
0.0958201569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002783_280 |
0.1015415791 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 7 |
Hb_000297_140 |
0.1022906713 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 8 |
Hb_000922_400 |
0.1063260597 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 9 |
Hb_006788_110 |
0.1073585418 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 10 |
Hb_000984_130 |
0.1077826991 |
- |
- |
tubulin [Ornithogalum longebracteatum] |
| 11 |
Hb_000140_320 |
0.1110846802 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 12 |
Hb_000103_070 |
0.1170235385 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 13 |
Hb_053264_010 |
0.117094483 |
- |
- |
beta glucosidase [Hevea brasiliensis] |
| 14 |
Hb_002471_130 |
0.1218811884 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 3 [Jatropha curcas] |
| 15 |
Hb_000878_050 |
0.1224094335 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 16 |
Hb_000820_110 |
0.1235224424 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 17 |
Hb_000103_450 |
0.124823873 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 18 |
Hb_003097_160 |
0.1258127304 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein sel-10 [Jatropha curcas] |
| 19 |
Hb_010962_020 |
0.1262138168 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_0815310 [Ricinus communis] |
| 20 |
Hb_004435_020 |
0.1264435125 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |