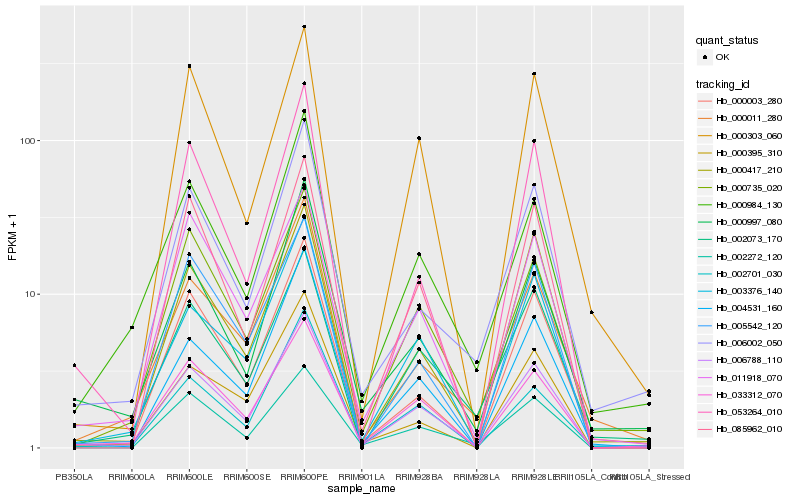
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006788_110 |
0.0 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 2 |
Hb_033312_070 |
0.071345603 |
- |
- |
hypothetical protein POPTR_0017s02960g [Populus trichocarpa] |
| 3 |
Hb_003376_140 |
0.0768123661 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 4 |
Hb_000735_020 |
0.0948547775 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 5 |
Hb_004531_160 |
0.099053409 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000984_130 |
0.1045462168 |
- |
- |
tubulin [Ornithogalum longebracteatum] |
| 7 |
Hb_011918_070 |
0.1051594851 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 8 |
Hb_002073_170 |
0.1063116889 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g27190 [Jatropha curcas] |
| 9 |
Hb_002272_120 |
0.1064207651 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 10 |
Hb_085962_010 |
0.1069598467 |
- |
- |
hypothetical protein POPTR_0017s11380g [Populus trichocarpa] |
| 11 |
Hb_006002_050 |
0.1073585418 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 12 |
Hb_000003_280 |
0.1077318544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000997_080 |
0.1089227931 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002701_030 |
0.1106063218 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 15 |
Hb_000417_210 |
0.1107637795 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000395_310 |
0.1108293653 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 17 |
Hb_005542_120 |
0.1109253189 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040 [Jatropha curcas] |
| 18 |
Hb_000011_280 |
0.1118657917 |
- |
- |
pectinesterase family protein [Populus trichocarpa] |
| 19 |
Hb_053264_010 |
0.1123654222 |
- |
- |
beta glucosidase [Hevea brasiliensis] |
| 20 |
Hb_000303_060 |
0.1148590281 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |