| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
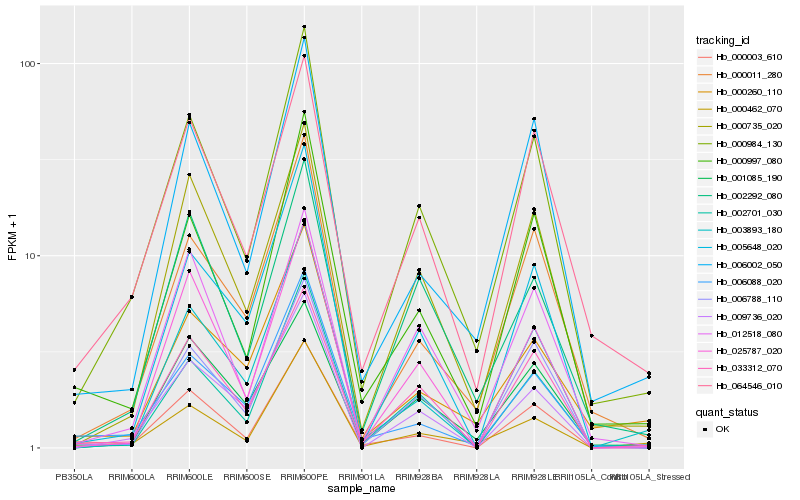
| 1 |
Hb_000984_130 |
0.0 |
- |
- |
tubulin [Ornithogalum longebracteatum] |
| 2 |
Hb_000997_080 |
0.0936117609 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000011_280 |
0.0960886116 |
- |
- |
pectinesterase family protein [Populus trichocarpa] |
| 4 |
Hb_064546_010 |
0.0982641444 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 5 |
Hb_000735_020 |
0.1023132113 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 6 |
Hb_006788_110 |
0.1045462168 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 7 |
Hb_002701_030 |
0.1058289311 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 8 |
Hb_006002_050 |
0.1077826991 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 9 |
Hb_000462_070 |
0.1111253797 |
- |
- |
PREDICTED: probable pectinesterase 15 [Jatropha curcas] |
| 10 |
Hb_002292_080 |
0.1134028055 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 11 |
Hb_012518_080 |
0.1174224691 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 12 |
Hb_033312_070 |
0.1195411366 |
- |
- |
hypothetical protein POPTR_0017s02960g [Populus trichocarpa] |
| 13 |
Hb_000003_610 |
0.1244268738 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase [Jatropha curcas] |
| 14 |
Hb_006088_020 |
0.1265782816 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 15 |
Hb_009736_020 |
0.1278126472 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG13 homolog [Jatropha curcas] |
| 16 |
Hb_001085_190 |
0.131344514 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 17 |
Hb_000260_110 |
0.1314436642 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 18 |
Hb_005648_020 |
0.1324992404 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 19 |
Hb_025787_020 |
0.1331936397 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_003893_180 |
0.1334555307 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |