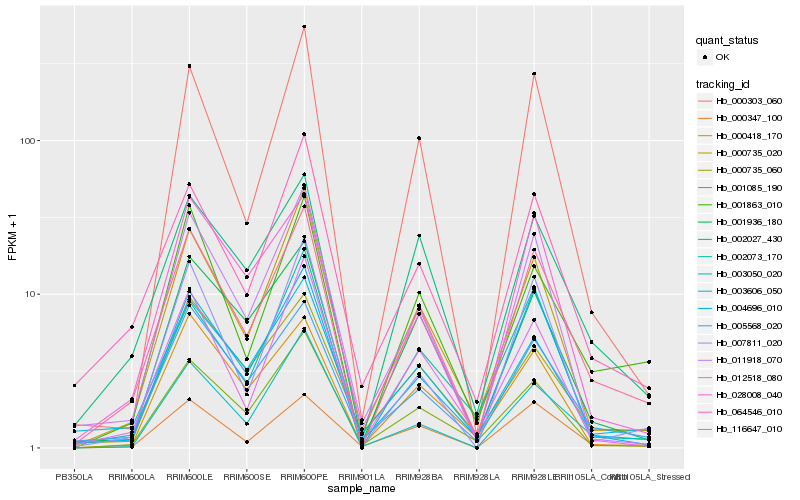
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116647_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005568_020 |
0.1024596654 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002027_430 |
0.1056197999 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 4 |
Hb_001863_010 |
0.1154928154 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 5 |
Hb_000735_020 |
0.1158949773 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 6 |
Hb_064546_010 |
0.1160727361 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 7 |
Hb_001085_190 |
0.120108622 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 8 |
Hb_003050_020 |
0.1201152837 |
- |
- |
glycosyl hydrolase family 17 family protein [Populus trichocarpa] |
| 9 |
Hb_004696_010 |
0.1212905456 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 10 |
Hb_000347_100 |
0.121716731 |
transcription factor |
TF Family: C3H |
hypothetical protein RCOM_0684740 [Ricinus communis] |
| 11 |
Hb_003606_050 |
0.122894147 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |
| 12 |
Hb_011918_070 |
0.1243347173 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 13 |
Hb_001936_180 |
0.1276173041 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 14 |
Hb_000303_060 |
0.1285905197 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_012518_080 |
0.1302846237 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 16 |
Hb_007811_020 |
0.1335778927 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 17 |
Hb_002073_170 |
0.1340692409 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g27190 [Jatropha curcas] |
| 18 |
Hb_028008_040 |
0.1364977335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000735_060 |
0.1368961466 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 20 |
Hb_000418_170 |
0.1392884423 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |