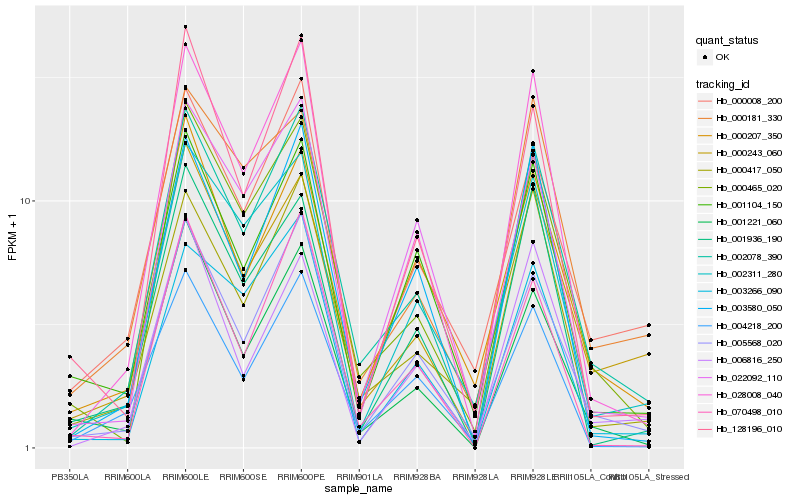
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_390 |
0.0 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 2 |
Hb_005568_020 |
0.08363381 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000465_020 |
0.0911439387 |
- |
- |
PREDICTED: uncharacterized protein LOC105632280 [Jatropha curcas] |
| 4 |
Hb_028008_040 |
0.0917752992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000243_060 |
0.1035499426 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 6 |
Hb_000417_050 |
0.1061286018 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 7 |
Hb_002311_280 |
0.1132602329 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 8 |
Hb_003580_050 |
0.1150077506 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 9 |
Hb_001221_060 |
0.1182548426 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 10 |
Hb_003266_090 |
0.1210287844 |
- |
- |
PREDICTED: basic 7S globulin 2-like [Jatropha curcas] |
| 11 |
Hb_000008_200 |
0.1220254927 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 12 |
Hb_022092_110 |
0.1248345866 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 13 |
Hb_000181_330 |
0.1251377818 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 14 |
Hb_000207_350 |
0.1252903236 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 15 |
Hb_001104_150 |
0.1280456085 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 16 |
Hb_004218_200 |
0.1281787142 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 17 |
Hb_070498_010 |
0.1294704651 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 18 |
Hb_001936_190 |
0.1299445004 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 19 |
Hb_006816_250 |
0.1323166235 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 20 |
Hb_128196_010 |
0.133572167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |