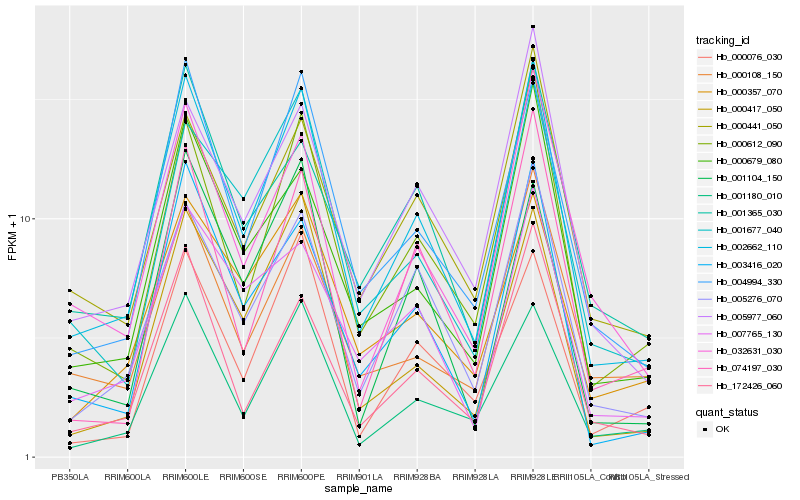
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002662_110 |
0.0 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 2 |
Hb_004994_330 |
0.0864504101 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_005276_070 |
0.0900020893 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 4 |
Hb_000612_090 |
0.0940273345 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 5 |
Hb_003416_020 |
0.0985670369 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 6 |
Hb_000679_080 |
0.1027527663 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_032631_030 |
0.1031064272 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 8 |
Hb_001180_010 |
0.1041143735 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 9 |
Hb_000417_050 |
0.1089949791 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 10 |
Hb_172426_060 |
0.1100234761 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 11 |
Hb_001677_040 |
0.1146400727 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 12 |
Hb_005977_060 |
0.1146805687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007765_130 |
0.1182873885 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000108_150 |
0.1201307993 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 15 |
Hb_001365_030 |
0.1201703156 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 16 |
Hb_000076_030 |
0.1207657738 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000441_050 |
0.1212306553 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 18 |
Hb_000357_070 |
0.1218434413 |
- |
- |
- |
| 19 |
Hb_074197_030 |
0.1228028497 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_001104_150 |
0.1228435085 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |