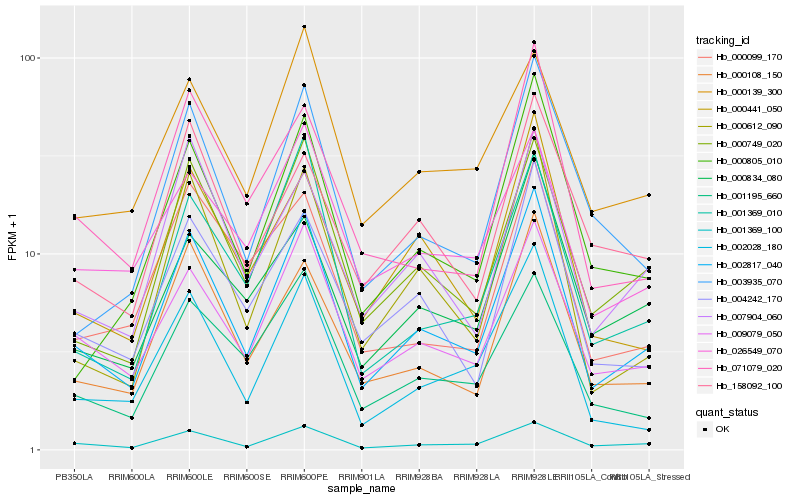
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002817_040 |
0.0 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 2 |
Hb_001369_100 |
0.0903655908 |
- |
- |
hypothetical protein JCGZ_26280 [Jatropha curcas] |
| 3 |
Hb_009079_050 |
0.0905472174 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 4 |
Hb_000749_020 |
0.0936770539 |
- |
- |
Nucleotide-diphospho-sugar transferases superfamily protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_000108_150 |
0.0999454814 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 6 |
Hb_000612_090 |
0.1037017799 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 7 |
Hb_002028_180 |
0.1098053493 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 8 |
Hb_004242_170 |
0.110809314 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 9 |
Hb_000441_050 |
0.1145527794 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 10 |
Hb_001195_660 |
0.1163758283 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 11 |
Hb_007904_060 |
0.119348919 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 12 |
Hb_000805_010 |
0.1199228839 |
- |
- |
PREDICTED: uncharacterized protein LOC105629931 [Jatropha curcas] |
| 13 |
Hb_003935_070 |
0.1215396066 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 14 |
Hb_071079_020 |
0.1242924463 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 15 |
Hb_001369_010 |
0.1244672206 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 16 |
Hb_000834_080 |
0.1262952747 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 17 |
Hb_026549_070 |
0.1296906097 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 18 |
Hb_000139_300 |
0.131752722 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 19 |
Hb_158092_100 |
0.1336599476 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 20 |
Hb_000099_170 |
0.1350487086 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |